Abstract
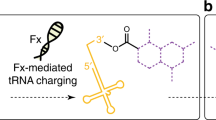
Genetic code reprogramming is a method for the reassignment of arbitrary codons from proteinogenic amino acids to nonproteinogenic ones; thus, specific sequences of nonstandard peptides can be ribosomally expressed according to their mRNA templates. Here we describe a protocol that facilitates genetic code reprogramming using flexizymes integrated with a custom-made in vitro translation apparatus, referred to as the flexible in vitro translation (FIT) system. Flexizymes are flexible tRNA acylation ribozymes that enable the preparation of a diverse array of nonproteinogenic acyl-tRNAs. These acyl-tRNAs read vacant codons created in the FIT system, yielding the desired nonstandard peptides with diverse exotic structures, such as N-methyl amino acids, D-amino acids and physiologically stable macrocyclic scaffolds. The facility of the protocol allows a wide variety of applications in the synthesis of new classes of nonstandard peptides with biological functions. Preparation of flexizymes and tRNA used for genetic code reprogramming, optimization of flexizyme reaction conditions and expression of nonstandard peptides using the FIT system can be completed by one person in ∼1 week. However, once the flexizymes and tRNAs are in hand and reaction conditions are fixed, synthesis of acyl-tRNAs and peptide expression is generally completed in 1 d, and alteration of a peptide sequence can be achieved by simply changing the corresponding mRNA template.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Forster, A.C. et al. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc. Natl. Acad. Sci. USA 100, 6353–6357 (2003).
Josephson, K., Hartman, M.C. & Szostak, J.W. Ribosomal synthesis of unnatural peptides. J. Am. Chem. Soc. 127, 11727–11735 (2005).
Murakami, H., Ohta, A., Ashigai, H. & Suga, H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods 3, 357–359 (2006).
Hecht, S.M., Alford, B.L., Kuroda, Y. & Kitano, S. 'Chemical aminoacylation' of tRNA's. J. Biol. Chem. 253, 4517–4520 (1978).
Wang, L., Xie, J. & Schultz, P.G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 35, 225–249 (2006).
Noren, C.J., Anthony-Cahill, S.J., Griffith, M.C. & Schultz, P.G. A general method for site-specific incorporation of unnatural amino acids into proteins. Science 244, 182–188 (1989).
Hohsaka, T., Ashizuka, Y., Murakami, H. & Sisido, M. Incorporation of nonnatural amino acids into streptavidin through in vitro frame-shift suppression. J. Am. Chem. Soc. 118, 9778–9779 (1996).
Cropp, T.A., Anderson, J.C. & Chin, J.W. Reprogramming the amino-acid substrate specificity of orthogonal aminoacyl-tRNA synthetases to expand the genetic code of eukaryotic cells. Nat. Protoc. 2, 2590–2600 (2007).
Murakami, H., Saito, H. & Suga, H. A versatile tRNA aminoacylation catalyst based on RNA. Chem. Biol. 10, 655–662 (2003).
Niwa, N., Yamagishi, Y., Murakami, H. & Suga, H. A flexizyme that selectively charges amino acids activated by a water-friendly leaving group. Bioorg. Med. Chem. Lett. 19, 3892–3894 (2009).
Sako, Y., Morimoto, J., Murakami, H. & Suga, H. Ribosomal synthesis of bicyclic peptides via two orthogonal inter-side-chain reactions. J. Am. Chem. Soc. 130, 7232–7234 (2008).
Goto, Y., Murakami, H. & Suga, H. Initiating translation with D-amino acids. RNA 14, 1390–1398 (2008).
Goto, Y. et al. Reprogramming the translation initiation for the synthesis of physiologically stable cyclic peptides. ACS Chem. Biol. 3, 120–129 (2008).
Kawakami, T., Murakami, H. & Suga, H. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem. Biol. 15, 32–42 (2008).
Kawakami, T., Murakami, H. & Suga, H. Ribosomal synthesis of polypeptoids and peptoid-peptide hybrids. J. Am. Chem. Soc. 130, 16861–16863 (2008).
Ohta, A., Murakami, H., Higashimura, E. & Suga, H. Synthesis of polyester by means of genetic code reprogramming. Chem. Biol. 14, 1315–1322 (2007).
Goto, Y. & Suga, H. Translation initiation with initiator tRNA charged with exotic peptides. J. Am. Chem. Soc. 131, 5040–5041 (2009).
Xiao, H., Murakami, H., Suga, H. & Ferre-D'Amare, A.R. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature 454, 358–361 (2008).
Kawakami, T. et al. Diverse backbone-cyclized peptides via codon reprogramming. Nat. Chem. Biol. 5, 888–890 (2009).
Hornbogen, T. & Zocher, R. Biosynthesis of N-methylated peptides in fungi. In Handbook of Industrial Mycology, Mycology Series v. 22 (ed., Zhiqiang, A.) 449–477 (Marcel Dekker, 2005).
Patch, J.A. & Barron, A.E. Mimicry of bioactive peptides via non-natural, sequence-specific peptidomimetic oligomers. Curr. Opin. Chem. Biol. 6, 872–877 (2002).
Sagan, S., Karoyan, P., Lequin, O., Chassaing, G. & Lavielle, S. N- and C alpha-methylation in biologically active peptides: synthesis, structural and functional aspects. Curr. Med. Chem. 11, 2799–2822 (2004).
Kwon, Y.U. & Kodadek, T. Quantitative evaluation of the relative cell permeability of peptoids and peptides. J. Am. Chem. Soc. 129, 1508–1509 (2007).
Sako, Y., Goto, Y., Murakami, H. & Suga, H. Ribosomal synthesis of peptidase-resistant peptides closed by a nonreducible inter-side-chain bond. ACS Chem. Biol. 3, 241–249 (2008).
Martinis, S.A. & Schimmel, P. Enzymatic aminoacylation of sequence-specific RNA minihelices and hybrid duplexes with methionine. Proc. Natl. Acad. Sci. USA 89, 65–69 (1992).
Putz, J. et al. Rapid selection of aminoacyl-tRNAs based on biotinylation of alpha-NH2 group of charged amino acids. Nucleic Acids Res. 25, 1862–1863 (1997).
Saito, H., Kourouklis, D. & Suga, H. An in vitro evolved precursor tRNA with aminoacylation activity. EMBO J. 20, 1797–1806 (2001).
Hopper, A.K. & Phizicky, E.M. tRNA transfers to the limelight. Genes Dev. 17, 162–180 (2003).
Goto, Y., Katoh, T. & Suga, H. Preparation of materials for flexizyme reactions and genetic code reprogramming. Protoc. Exchange. doi:10.1038/protex.2011.209 (2011).
Olins, P.O., Devine, C.S., Rangwala, S.H. & Kavka, K.S. The T7 phage gene 10 leader Rna, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene 73, 227–235 (1988).
Kung, H.F. et al. DNA-directed in vitro synthesis of beta-galactosidase. Studies with purified factors. J. Biol. Chem. 252, 6889–6894 (1977).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Charlton, A. & Zachariou, M. Immobilized metal ion affinity chromatography of histidine-tagged fusion proteins. Methods Mol. Biol. 421, 137–149 (2008).
Block, H. et al. Immobilized-metal affinity chromatography (IMAC): a review. Meth. Enzymol. 463, 439–473 (2009).
Schagger, H. Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 (2006).
Shukla, A. & Majors, R.E. Micropipette tip-based sample preparation for bioanalysis. LCGC Eur. 18, 650 (2005).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Milligan, J.F., Groebe, D.R., Witherell, G.W. & Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 (1987).
Clemons, W.M. Jr. et al. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: purification, crystallization and structure determination. J. Mol. Biol. 310, 827–843 (2001).
Baggott, J.E. et al. Cofactor role for 10-formyldihydrofolic acid. Biochem. J. 308 (Part 3): 1031–1036 (1995).
Acknowledgements
We thank Y. Yamagishi for the discussion and proofreading. We thank H. Murakami and T. Kawakami for the contributions to the development of the methods presented in this study. We also thank P.C. Reid and C.J. Hipolito for proofreading. This work was supported by grants from the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (S) (16101007), Specially Promoted Research (21000005), a research and development projects of the Industrial Science and Technology Program in the New Energy and Industrial Technology Development Organization, and the World Class University project of the MEST and the NRF (R31-2008-000-10103-0) to H.S.; and grants from the Japan Society for the Promotion of Science Grants-in-Aid for Young Scientists (B) (22750145) to Y.G. and (B) (22710210) to T.K.
Author information
Authors and Affiliations
Contributions
Y.G. developed the methods presented in this study. Y.G. and T.K. validated the protocol, wrote the article and prepared the figures. H.S. supervised all the work and prepared the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
We disclosed a patent related to this technology when this manuscript was submitted. Note that the patent includes some technical advancements to the FIT system that are not yet ready for publication.
Rights and permissions
About this article
Cite this article
Goto, Y., Katoh, T. & Suga, H. Flexizymes for genetic code reprogramming. Nat Protoc 6, 779–790 (2011). https://doi.org/10.1038/nprot.2011.331
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.331
This article is cited by
-
Alternative therapeutic strategies to treat antibiotic-resistant pathogens
Nature Reviews Microbiology (2024)
-
Synthesis and applications of mirror-image proteins
Nature Reviews Chemistry (2023)
-
In vitro selection of macrocyclic peptide inhibitors containing cyclic γ2,4-amino acids targeting the SARS-CoV-2 main protease
Nature Chemistry (2023)
-
Cell-free Biosynthesis of Peptidomimetics
Biotechnology and Bioprocess Engineering (2023)
-
A mating mechanism to generate diversity for the Darwinian selection of DNA-encoded synthetic molecules
Nature Chemistry (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.