Abstract
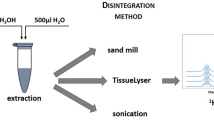
This protocol describes an analytical platform for the analysis of intra- and extracellular metabolites of microbial cells (yeast, filamentous fungi and bacteria) using gas chromatography–mass spectrometry (GC-MS). The protocol is subdivided into sampling, sample preparation, chemical derivatization of metabolites, GC-MS analysis and data processing and analysis. This protocol uses two robust quenching methods for microbial cultures, the first of which, cold glycerol-saline quenching, causes reduced leakage of intracellular metabolites, thus allowing a more reliable separation of intra- and extracellular metabolites with simultaneous stopping of cell metabolism. The second, fast filtration, is specifically designed for quenching filamentous micro-organisms. These sampling techniques are combined with an easy sample-preparation procedure and a fast chemical derivatization reaction using methyl chloroformate. This reaction takes place at room temperature, in aqueous medium, and is less prone to matrix effect compared with other derivatizations. This protocol takes an average of 10 d to complete and enables the simultaneous analysis of hundreds of metabolites from the central carbon metabolism (amino and nonamino organic acids, phosphorylated organic acids and fatty acid intermediates) using an in-house MS library and a data analysis pipeline consisting of two free software programs (Automated Mass Deconvolution and Identification System (AMDIS) and R).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout














Similar content being viewed by others
References
Castoldi, M., Schmidt, S., Benes, V., Hentze, M.W. & Muckenthaler, M.U. miChip: an array-based method for microRNA expression profiling using locked nucleic acid capture probes. Nat. Protoc. 3, 321–329 (2008).
Harsha, H.C., Molina, H. & Pandey, A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat. Protoc. 3, 505–516 (2008).
Villas-Bôas, S.G., Mas, S., Åkesson, M., Smedsgaard, J. & Nielsen, J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 24, 613–646 (2005).
Kell, D.B. Metabolomics and systems biology: making sense of the soup. Curr. Opin. Microbiol. 7, 296–307 (2004).
Dunn, W.B., Bailey, N.J.C. & Johnson, H.E. Measuring the metabolome: current analytical technologies. Analyst 130, 606–625 (2005).
Wang, Q., Wu, C., Chen, T., Chen, X. & Zhao, X. Integrating metabolomics into systems biology framework to exploit metabolic complexity: strategies and applications in microorganisms. Appl. Microbiol. Biotechnol. 70, 151–161 (2006).
Villas-Bôas, S.G. & Bruheim, P. Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal. Biochem. 370, 87–97 (2007).
van der Werf, M.J., Overkamp, K.M., Muilwijk, B., Coulier, L. & Hankemeier, T. Microbial metabolomics: toward a platform with full metabolome coverage. Anal. Biochem. 370, 17–25 (2007).
Schauer, N. et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579, 1332–1337 (2005).
Villas-Bôas, S.G. Sampling and sample preparation. In Metabolome Analysis: An Introduction (eds. Villas-Bôas, S.G., Roessner, U., Hansen, M.A.E., Smedsgaard, J. & Nielsen, J) 39–82 (John Wiley & Sons, Hoboken, New Jersey, USA, 2007).
Lin, Y., Schiavo, S., Orjala, J., Vouros, P. & Kautz, R. Microscale LC-MS-NMR platform applied to the identification of active cyanobacterial metabolites. Anal. Chem. 80, 8045–8054 (2008).
Bundy, J.G., Willey, T.L., Castell, R.S., Ellar, D.J. & Brindle, K.M. Discrimination of pathogenic clinical isolates and laboratory strains of Bacillus cereus by NMR-based metabolomic profiling. FEMS Microbiol. Lett. 242, 127–136 (2005).
Koek, M.M., Muilwijk, B., van der Werf, M.J. & Hankemeier, T. Microbial metabolomics with gas chromatography/mass spectrometry. Anal. Chem. 78, 1272–1281 (2006).
Roessner, U. et al. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant. J. 23, 131–142 (2000).
Smedsgaard, J. Analytical tools. In Metabolome Analysis: An Introduction (eds. Villas-Bôas, S.G., Roessner, U., Hansen, M.A.E., Smedsgaard, J. & Nielsen, J) 83–145 (John Wiley & Sons, Hoboken, New Jersey, USA, 2007).
Villas-Bôas, S.G., Noel, S., Lane, G.A., Attwood, G. & Cookson, A. Extracellular metabolomics: a metabolic footprinting approach to assess fiber degradation in complex media. Anal. Biochem. 349, 297–305 (2006).
Kanani, H.H. & Klapa, M.I. Data correlation strategy for metabolomics analysis using gas chromatography-mass spectrometry. Metab. Eng. 9, 39–51 (2007).
Winder, C.L. et al. Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal. Chem. 80, 2939–2948 (2008).
Villas-Bôas, S.G., Delicado, D.G., Åkesson, M. & Nielsen, J. Simultaneous analysis of amino and nonamino organic acids as methyl chloroformate derivatives using gas chromatography-mass spectrometry. Anal. Biochem. 322, 134–138 (2003).
Sumner, L.W., Mendes, P. & Dixon, R.A. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62, 817–836 (2003).
Park, S.J. et al. Global physiological understanding and metabolic engineering of microorganisms based on omics studies. Appl. Microbiol. Biotechnol. 68, 567–579 (2005).
Bro, C. & Nielsen, J. Impact of 'ome' analyses on inverse metabolic engineering. Metab. Eng. 6, 204–211 (2004).
Lee, S.Y., Lee, D. & Kim, T.Y. Systems biotechnology for strain improvement. Trends Biotechnol. 23, 349–358 (2005).
Styczynski, M.P. et al. Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Anal. Chem. 79, 966–973 (2007).
Mapelli, V., Olsson, L. & Nielsen, J. Metabolic footprinting in microbiology: methods and applications in functional genomics and biotechnology. Trends Biotechnol. 26, 490–497 (2008).
Villas-Bôas, S.G., Åkesson, M. & Nielsen, J. Biosynthesis of glyoxylate from glycine in Saccharomyces cerevisiae. FEMS Yeast Res. 5, 703–709 (2005).
Villas-Bôas, S.G., Moxley, J.F., Åkesson, M., Stephanopoulos, G. & Nielsen, J. High-throughput metabolic state analysis: the missing link in integrated functional genomics of yeasts. Biochem. J. 388, 669–677 (2005).
Büscher, J.M., Czernik, D., Ewald, J.C., Sauer, U. & Zamboni, N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal. Chem. 81, 2135–2143 (2009).
Ewald, J.C., Heux, S. & Zamboni, N. High-throughput quantitative metabolomics: workflow for cultivation, quenching, and analysis of yeast in a multiwell format. Anal. Chem. 81, 3623–3629 (2009).
Villas-Bôas, S.G., Højer-Pedersen, J., Åkesson, M., Smedsgaard, J. & Nielsen, J. Global metabolite analysis of yeasts: evaluation of sample preparation methods. Yeast 22, 1155–1169 (2005).
Mas, S., Villas-Bôas, S.G., Hansen, M.E., Åkesson, M. & Nielsen, J. A comparison of direct infusion MS with GC-MS for metabolic footprinting of yeast mutants. Biotechnol. Bioeng. 96, 1014–1022 (2007).
Panagiotou, G., Villas-Bôas, S.G., Christakopoulos, P., Nielsen, J. & Olsson, L. Intracellular metabolite profiling of Fusarium oxysporum converting glucose to ethanol. J. Biotechnol. 115, 425–434 (2005).
Villas-Bôas, S.G. et al. Phenotypic characterization of transposon-inserted mutants of Clostridium proteoclasticum B316T using extracellular metabolomics. J. Biotechnol. 134, 55–63 (2008).
Mashego, M.R. et al. MIRACLE: Mass isotopomer ratio analysis of U-13C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol. Bioeng. 85, 620–628 (2004).
Villas-Bôas, S.G., Koulman, A. & Lane, G.A. Analytical methods from the perspective of method standardization. In Topics in Current Genetics—Metabolomics (eds. Nielsen, J. & Jewett, M.C.) 11–52 (Springer-Verlag, Berlin Heidelberg, Germany, 2007).
Bolten, C.J., Kiefer, P., Letisse, F., Portais, J.C. & Wittmann, C. Sampling for metabolome analysis of microorganisms. Anal. Chem. 79, 3843–3849 (2007).
Brauer, M.J. et al. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA 103, 19302–19307 (2006).
Wittmann, C., Krömer, J.O., Kiefer, P., Binz, T. & Heinzle, E. Impact of the cold shock phenomenon on quantification of intracellular metabolites in bacteria. Anal. Biochem. 327, 135–139 (2004).
Förster, J., Famili, I., Fu, P., Palsson, B.Ø. & Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13, 244–253 (2003).
Stein, S.E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry. J. Am. Soc. Mass Spectrom. 10, 770–781 (1999).
Freisleben, A., Schieberle, P. & Rychlik, M. Specific and sensitive quantification of folate vitamers by stable isotope dilution assays using high-performance liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 376, 149–156 (2003).
Bennett, B.D., Yuan, J., Kimball, E.H. & Rabinowitz, J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 3, 1299–1311 (2008).
Smith, C.A., Want, E.J., O'Maille, G., Abagyan, R. & Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Kopka, J. et al. GMD@CSB.DB: the Golm metabolome database. Bioinformatics 21, 1635–1638 (2005).
Tayrac, M., Lê, S., Aubry, M., Mosser, J. & Husson, F. Simultaneous analysis of distinct Omics data sets with integration of biological knowledge: multiple factor analysis approach. BMC Genomics 10, 32 (2009).
Mendes, P., Camacho, D. & de la Fuente, A. Modelling and simulation for metabolomics data analysis. Biochem. Soc. Trans. 33, 1427–1429 (2005).
Jansen, J.J., Hoefsloot, H.C.J., Boelens, H.F.M., van der Greef, J. & Smilde, A.K. Analysis of longitudinal metabolomics data. Bioinformatics 20, 2438–2446 (2004).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Devantier, R., Scheithauer, B., Villas-Bôas, S.G., Pedersen, S. & Olsson, L. Metabolite profiling for analysis of yeast stress response during very high gravity ethanol fermentations. Biotechnol. Bioeng. 90, 703–714 (2005).
Moxley, J.F. et al. Linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator Gcn4p. Proc. Natl Acad. Sci. USA 106, 6477–6482 (2009).
Acknowledgements
We thank the contribution of all people involved at different stages of the development of the methods summarized here, in particular, J. Nielsen, M. Åkesson, J.F. Moxley, P. Bruheim and G. Lane. We also thank the following institutions: Technical University of Denmark, The Norwegian University of Science and Technology, SINTEF Materials and Chemistry and AgResearch Limited.
Author information
Authors and Affiliations
Contributions
K.S. co-wrote the paper and optimized the protocol and videos; R.B.M.A. co-wrote the paper and generated the scripts for data mining, figures and videos; J.V.H. edited the MCF library and videos; and S.V.-B. supervised writing and developed the analytical platform.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data 1
MCFMS.CID file from MCF MS Library. Our in house library consists of two files: MCFMS.CID and MCFMS.MSL (supplemented material 2). Box 1 presents all steps and necessary instructions to load our in-house library in AMDIS. Download the files and follow the instruction in Box 1. (TXT 141 kb)
Supplementary Data 2
MCFMS.MSL file from MCF MS Library. Our in house library consists of two files: MCFMS.CID (supplemented material 1) and MCFMS.MSL. Box 1 presents all steps and necessary instructions to load our in-house library in AMDIS. Download the files and follow the instruction in Box 1. (TXT 141 kb)
Supplementary Data 3
Step 21(B) (i). Our in-house script consists of one R scrip file called ‘mining_script’ and all requirements and instructions to use it are described in the first rows of the script. (TXT 47 kb)
Supplementary Data 4
Step 21(B) (i). The reference ion library consists of one comma separated file (.csv) called ‘ref_ion_library.csv’. This file is complementary to the in-house script (mining_script) and both files have to be stored in the same folder. (CSV 15 kb)
Supplementary Video 1
Step 2 (A) (i), (ii). Quenching using cold glycerol-saline. Rapid transfer microbial culture suspension to a cold solution of glycerol-saline at −23°C. Note that the volume of sample transferred can be roughly controlled by level marks in the sampling flasks. This is not accurate and, therefore, the quantification of biomass in each sample must be carried out after extraction. (WMV 2559 kb)
Supplementary Video 2
Step 14 (A) (iii)-(v). MCF reaction after the samples have been resuspended in NaOH and mixed with methanol and pyridine. Although it is not clear in this video, the vortex agitation must be “vigorous”. (WMV 3788 kb)
Supplementary Video 3
Step 14 (A) (vii). Removal of aqueous phase. The top aqueous layer should be totally removed. (WMV 1043 kb)
Supplementary Video 4
Step 14 (A) (ix). End of derivatization. Sodium sulphate-dried chloroform solution containing the MCF derivatives is transferred to the GC-MS vial. Avoid transferring granules of sodium sulphate together with the sample. (WMV 2399 kb)
Rights and permissions
About this article
Cite this article
Smart, K., Aggio, R., Van Houtte, J. et al. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography–mass spectrometry. Nat Protoc 5, 1709–1729 (2010). https://doi.org/10.1038/nprot.2010.108
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.108
This article is cited by
-
RBM15 suppresses hepatic insulin sensitivity of offspring of gestational diabetes mellitus mice via m6A-mediated regulation of CLDN4
Molecular Medicine (2023)
-
Segmental hair metabolomics analysis in pregnant women with pregnancy complications
Metabolomics (2023)
-
Chemical Compositions of Fruit and Vegetable Pomaces from the Beverage Industries
Waste and Biomass Valorization (2023)
-
Metabolite profiling of abalone (Haliotis iris) energy metabolism: a Chatham Islands case study
Metabolomics (2022)
-
Plasma nervonic acid levels were negatively associated with attention levels in community-living older adults in New Zealand
Metabolomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.