Abstract
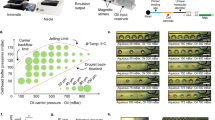
Single molecule-based protocols have been gaining popularity as a way to visualize DNA replication at the global genomic- and locus-specific levels. These protocols take advantage of the ability of many organisms to incorporate nucleoside analogs during DNA replication, together with a method to display stretched DNA on glass for immunostaining and microscopy. We describe here a microfluidic platform that can be used to stretch and to capture labeled DNA molecules for replication analyses. This platform consists of parallel arrays of three-sided, 3- or 4-μm high, variable-width capillary channels fabricated from polydimethylsiloxane by conventional soft lithography, and of silane-modified glass coverslips to reversibly seal the open side of the channels. Capillary tension in these microchannels facilitates DNA loading, stretching and glass coverslip deposition from microliter-scale DNA samples. The simplicity and extensibility of this platform should facilitate DNA replication analyses using small samples from a variety of biological and clinical sources.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Alberts, B. DNA replication and recombination. Nature 421, 431–435 (2003).
Kelly, T. & Stillman, B. Duplication of DNA in eukaryotic cells. In DNA Replication and Human Disease (ed. DePamphilis, M.L.) 1–29 (Cold Spring Harbor Press, Cold Spring Harbor, New York, 2006).
Loeb, L.A. & Monnat, R.J. DNA polymerases and human disease. Nat. Rev. Genet. 9, 594–604 (2008).
McCulloch, S.D. & Kunkel, T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18, 148–161 (2008).
Yang, W. & Woodgate, R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. USA 104, 15591–15598 (2007).
Gilson, E. & Geli, V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838 (2007).
Bianchi, A. & Shore, D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol. Cell 31, 153–165 (2008).
Kornberg, A. & Baker, T.A. DNA Replication (W.H. Freeman, New York, 1991).
Pommier, Y. & Diasio, R.B. Pharmacologic agents that target DNA replication. In DNA Replication and Human Disease (ed. DePamphilis, M.L.) 519–546 (Cold Spring Harbor Press, Cold Spring Harbor, New York, 2006).
Berdis, A.J. DNA polymerases as therapeutic targets. Biochemistry 47, 8253–8260 (2008).
Vengrova, S. & Dalgaard, J.Z. Eds. DNA Replication Methods and Protocols (Humana Press, New York, 2009).
Manders, E.M., Stap, J., Brakenhoff, G.J., van Driel, R. & Aten, J.A. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 103, 857–862 (1992).
Jackson, D.A. & Pombo, A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–95 (1998).
Huberman, J.A. & Riggs, A.D. Autoradiography of chromosomal DNA fibers from Chinese hamster cells. Proc. Natl. Acad. Sci. USA 55, 599–606 (1966).
Cairns, J. The chromosome of Escherichia coli . In Cold Spring Harbor Symposia on Quantitative Biology Vol. 28 43–46 (Cold Spring Harbor Press, Cold Spring Harbor, New York, 1963).
Bensimon, A. et al. Alignment and sensitive detection of DNA by a moving interface. Science 265, 2096–2098 (1994).
Yokota, H. et al. A new method for straightening DNA molecules for optical restriction mapping. Nucleic Acids Res. 25, 1064–1070 (1997).
Herrick, J. & Bensimon, A. Single molecule analysis of DNA replication. Biochimie 81, 859–871 (1999).
Norio, P. & Schildkraut, C.L. Visualization of DNA replication on individual Epstein–Barr virus episomes. Science 294, 2361–2364 (2001).
Sidorova, J.M., Li, N., Folch, A. & Monnat, R.J. Jr. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 7, 796–807 (2008).
Dimalanta, E.T. et al. A microfluidic system for large DNA molecule arrays. Anal. Chem. 76, 5293–5301 (2004).
Zhou, S., Herschleb, J. & Schwartz, D.C. A single molecule system for whole genome analysis. In New High Throughput Technologies for DNA Sequencing and Genomics (ed. Mitchelson, K.R.) 265–302 (Elsevier, Amsterdam, 2007).
Zhou, S. et al. Validation of rice genome sequence by optical mapping. BMC Genomics 8, 278 (2007).
Valouev, A., Schwartz, D., Zhou, S. & Waterman, M.S. An algorithm for assembly of ordered restriction maps from single DNA molecules. Proc. Natl. Acad. Sci. USA 103, 15770–15775 (2006).
Kidd, J.M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
McDonald, J.C. et al. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 21, 27–40 (2000).
Beebe, D.J., Mensing, G.A. & Walker, G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 4, 261–286 (2002).
Tegenfeldt, J. et al. Micro- and nanofluidics for DNA analysis. Anal. Bioanal. Chem. 378, 1678–1692 (2004).
Atencia, J. & Beebe, D.J. Controlled microfluidic interfaces. Nature 437, 648–655 (2005).
Whitesides, G.M. The origins and the future of microfluidics. Nature 442, 368–373 (2006).
Herschleb, J., Ananiev, G. & Schwartz, D.C. Pulsed-field gel electrophoresis. Nat. Protoc. 2, 677–684 (2007).
Lundin, C. et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33, 3799–3811 (2005).
Acknowledgements
This work was supported by the NIH Seattle Cancer and Aging Program Grant P20 CA103728Subaward 000618167 to J.S., an NHGRI R01 (HG000225) to D.C.S., an R21/R33 EB0003307 award to A.F. and NCI PO1 CA88752 to R.J.M. Jr. We also thank Dhalia Dhingra and Paolo Norio for advice during early phases of this work, Amelia Gallaher for technical assistance and Alden Hackmann for graphics support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sidorova, J., Li, N., Schwartz, D. et al. Microfluidic-assisted analysis of replicating DNA molecules. Nat Protoc 4, 849–861 (2009). https://doi.org/10.1038/nprot.2009.54
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.54
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.