Abstract
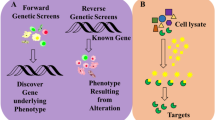
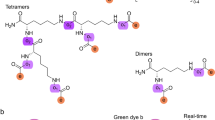
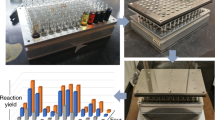
One of the most fundamental properties of an enzyme is its selectivity, a property that has proved highly challenging to understand. Recent developments offer methodologies to rapidly establish activity-dependent profiles of enzymes. In this protocol, we describe methods to elucidate inhibitor fingerprints of enzymes. By taking advantage of well-defined small-molecule inhibitor libraries and the screening throughput offered from microplate and microarray platforms, we provide step-by-step application of the methodology toward the global characterization of metalloproteases, an important class of enzymes involved in numerous diseases and cellular processes. The same strategy is nonetheless applicable to virtually any given enzyme class, provided suitable experimental design and chemical inhibitor libraries are carefully implemented. We are able to routinely fingerprint as many as 2,000 independent enzyme interactions on the microplate platform within a span of ∼7 h; however, the same throughput is attained within 5 h on the microarray platform.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Creighton, T.E. Proteins: Structure and Molecular Properties 2nd edn. (Freeman, New York, 1993).
Sun, H., Chattopadhaya, S., Wang, J. & Yao, S.Q. Recent development in microarray-based enzyme assays: from functional annotation to substrate/inhibitor fingerprinting. Anal. Bioanal. Chem. 386, 416–426 (2006).
Chang, H.T. et al. Reinforced merging methodology for mapping unique peptide motifs in members of protein families. BMC Bioinformatics 7, 38 (2006).
Mager, P.P., Weber, A. & Illes, P. Bridging the gap between structural bioinformatics and receptor research: the membrane-embedded, ligand-gated, P2X glycoprotein receptor. Curr. Top. Med. Chem. 4, 1657–1705 (2004).
Sanchez, R. & Sali, A. Advances in comparative protein-structure modeling. Curr. Opin. Struct. Biol. 7, 206–214 (1997).
Uttamchandani, M., Liu, K., Panicker, R.C. & Yao, S.Q. Activity-based fingerprinting and inhibitor discovery of cysteine proteases in a microarray. Chem. Commun. 1518–1520 (2007).
Wang, J., Uttamchandani, M., Li, J., Hu, M. & Yao, S.Q. “Click” synthesis of small molecule probes for activity-based fingerprinting of matrix metalloproteases. Chem. Commun. 3783–3785 (2006).
Wang, J., Uttamchandani, M., Sun, L.P. & Yao, S.Q. Activity-based high-throughput profiling of metalloprotease inhibitors using small molecule microarrays. Chem. Commun. 717–719 (2006).
Srinivasan, R., Huang, X., Ng, S.L. & Yao, S.Q. Activity-based fingerprinting of proteases. ChemBioChem 7, 32–36 (2006).
Uttamchandani, M. et al. Inhibitor fingerprinting of matrix metalloproteases using a combinatorial peptide hydroxamate library. J. Am. Chem. Soc. 129, 7848–7858 (2007).
Choe, Y. et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 281, 12824–12832 (2006).
Debela, M. et al. Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J. Biol. Chem. 281, 25678–25688 (2006).
Harris, J.L. et al. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. USA 97, 7754–7759 (2000).
Xiao, X.Y. et al. Solid-phase combinatorial synthesis using MicroKan reactors, Rf tagging, and directed sorting. Biotechnol. Bioeng. 71, 44–50 (2000).
Overall, C.M. & Lopez-Otin, C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat. Rev. Cancer 2, 657–672 (2002).
Uttamchandani, M., Wang, J. & Yao, S.Q. Protein and small molecule microarrays: powerful tools for high-throughput proteomics. Mol. BioSyst. 2, 58–68 (2006).
Lutz, W. O-Triphenylmethylhydroxylamine (trityloxyamine), a useful O-protected form of hydroxylamine. J. Org. Chem. 36, 3835–3837 (1971).
Knight, C.G., Willenbrock, F. & Murphy, G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 296, 263–266 (1992).
Kaiser, E., Colescott, R.L., Bossinger, C.D. & Cook, P.I. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 34, 595–598 (1992).
Chattopadhaya, S., Tan, L.P. & Yao, S.Q. Strategies for site-specific protein biotinylation using in vitro, in vivo and cell-free systems: toward functional protein arrays. Nat. Protoc. 1, 2386–2398 (2006).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Lee, W., Li, J., Uttamchandani, M. et al. Inhibitor fingerprinting of metalloproteases using microplate and microarray platforms: an enabling technology in Catalomics. Nat Protoc 2, 2126–2138 (2007). https://doi.org/10.1038/nprot.2007.305
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.305
This article is cited by
-
Peptide microarrays for high-throughput studies of Ser/Thr phosphatases
Nature Protocols (2008)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.