Abstract
Protein glycosylation is a common post-translational modification and has been increasingly recognized as one of the most prominent biochemical alterations associated with malignant transformation and tumorigenesis. N-linked glycosylation is prevalent in proteins on the extracellular membrane, and many clinical biomarkers and therapeutic targets are glycoproteins. Here, we describe a protocol for solid-phase extraction of N-linked glycopeptides and subsequent identification of N-linked glycosylation sites (N-glycosites) by tandem mass spectrometry. The method oxidizes the carbohydrates in glycopeptides into aldehydes, which can be immobilized on a solid support. The N-linked glycopeptides are then optionally labeled with a stable isotope using deuterium-labeled succinic anhydride and the peptide moieties are released by peptide-N-glycosidase. In a single analysis, the method identifies hundreds of N-linked glycoproteins, the site(s) of N-linked glycosylation and the relative quantity of the identified glycopeptides.
Similar content being viewed by others
Introduction
Glycosylation has long been recognized as the most common protein post-translational modification. It affects protein function, such as protein localization, stability, enzymatic activity and protein–protein interactions1. Differential glycosylation is a major source of protein microheterogeneity. Glycosylation plays key roles in cell communication, signaling and cell adhesion. Changes in carbohydrates in cell surface and body fluid proteins have been demonstrated in cancer and other conditions and highlight the importance of this modification2,3. However, studies on protein glycosylation have been complicated by the diverse structure of protein glycans and the lack of effective tools to identify the protein glycosylation site and glycan structure. For example, human protein database contains thousands of predicted N-linked glycosylated proteins, however, before high-throughput methods for the identification of glycosites were developed in 2003, only 172 proteins were proven to be glycoproteins experimentally in the Protein Information Resources-Protein Sequence Database (http://pir.georgetown.edu/pirwww/search/textpsd.shtml)4.
Recently, two new methods for isolation and identification of N-linked glycopeptides in complex biological samples––solid-phase extraction of N-linked glycopeptides (SPEG) and glycopeptide capture using lectin-affinity column chromatography––have been reported4,5. In both instances, mass spectrometry is used after the isolation of the N-glycopeptides to identify the N-glycosites and quantify the relative abundance of glycopeptides using isotope-coded tags.
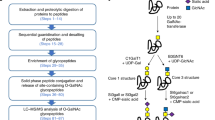
In the first method, glycoproteins are covalently conjugated to a solid support via hydrazide chemistry, and non-glycosylated peptides are removed with proteolysis before the release of N-linked glycopeptides from solid support5. In the modified method detailed in this protocol (Fig. 1), glycoproteins are first digested into peptides that contain both glycosylated peptides and non-glycosylated peptides, and the cis-diol groups of carbohydrates in glycopeptides are oxidized to aldehydes, which then form covalent hydrazone bonds with hydrazide groups immobilized on a solid support. Non-glycosylated peptides are washed away, whereas the glycosylated peptides remain on the solid support. For accurate quantitative analysis of N-linked glycopeptides using isotope labeling and mass spectrometry, the amino groups of the immobilized glycopeptides can be labeled with isotopic tags. For example, light (d0, containing no deuteriums) or heavy (d4, containing four deuteriums) forms of succinic anhydrides can be used to label the glycopeptides isolated from two biological samples. Last, the formerly N-linked glycosylated peptides are released from the solid phase using peptide-N-glycosidase F (PNGase F). PNGase F treatment also results in the conversion of the glycosylated asparagines to aspartic acids, thus generating a 1 U mass shift at the site of glycosylation, which is detectable using a high-accuracy mass spectrometer and is diagnostic for the glycosylation site. Therefore, in a single analysis, the method identifies N-linked glycosylated proteins, the site(s) of N-linked glycosylation and the relative quantity of the identified glycopeptides via stable isotope tagging.
Proteins are first proteolyzed into peptides. Glycosylated peptides are then oxidized and coupled to a solid support. Non-glycopeptides are removed by successive washes. The amino-termini of glycopeptides are labeled by succinic anhydride carrying either d0 or d4. N-linked glycopeptides are then released by PNGase F and analyzed by mass spectrometry.
In the second approach, glycopeptides are immobilized by lectin column-mediated affinity capture. After elution of glycopeptides from lectin column, peptide-N-glycosidase is then used to release the N- glycopeptides and the resulting peptides are subjected to mass spectrometry to identify the N-linked glycopeptides and sites of glycosylation4. For glycopeptide capture using hydrazide chemistry, peptides containing either N-linked or O-linked oligosaccharides are conjugated to a solid support covalently. Non-glycosylated peptides can be removed by extensive washing before the release of glycopeptides; therefore, the glycopeptides can be specifically enriched (over 90% enrichment6). The specific release of different types of glycopeptides from solid support can be achieved by varying glycosidases or chemicals. For glycopeptide capture using a lectin affinity column, glycopeptides containing certain glycan structures can be selectively enriched by using different types of lectin affinity chromatography. The specificity and reproducibility may not be easily controlled due to affinitive capture of glycopeptides.
Extracellular proteins, such as proteins expressed on the cell surface or secreted from the cell, are exposed to extracellular environments such as surrounding tissue, blood or other body fluids. Such proteins are thus the most easily accessible for diagnostic and therapeutic purposes. If present in easily accessible body fluids such as blood plasma or cerebrospinal fluid, they are also preferred candidates for protein biomarkers. However, the challenge faced by all proteomic methods for the analysis of body fluids is the peculiar properties of these samples. The proteome of most body fluids is complex, consisting minimally of tens of thousands of different protein species and exhibiting a high dynamic range in protein concentration. These proteomes are also dominated by a few highly abundant proteins7, and as a result, most proteomic analyses of body fluids detect only a limited number of highly abundant proteins. Although the dynamic range of protein abundances in tissues or cells is not as large as it is in body fluids, a similar situation is still observed: proteomic analyses detect only the most abundant protein subset, comprised mostly of structural intracellular proteins. Glycosylation, especially N-linked glycosylation, is a common modification of proteins that are exposed to an extracellular environment8. Therefore, selective isolation of N-linked glycopeptides enriches tissue and body fluid proteomes for extracellular proteins. In addition, the number of N-linked glycosylation sites in the human extracellular proteome is modest and is identifiable with current proteomic technology. For example, N-glycosites, which generally contain the N-X-S/T sequence motif (where X denotes any amino acid except proline)9, are found in just 3% of tryptic peptides from the entire human proteome; yet they represent the majority of extracellular proteins (over 70%) (see ref. 10). Therefore, targeted analysis of N-glycosites significantly increases the sensitivity for low-abundance glycoproteins by reducing the number of detectable peptides at each targeted mass range.
Although the analysis of N-linked glycopeptides reduces sample complexity and peptide redundancy and is therefore beneficial for achieving higher coverage of the proteome per analysis, it is also apparent that it leads to the loss of some, potentially important information. First, non-glycosylated proteins are transparent to this system. Second, the availability of a few N-glycosites per protein increases the challenge of identifying the corresponding protein. Third, tryptic peptides that are too short or too long to fall within the detection range of the mass spectrometer will not be identified, although this last limitation may be overcome, at least in part, by the use of proteases with cleavage specificities different from that of trypsin. Fourth, removal of oligosaccharides from the N-linked glycopeptides before their analysis only identifies the N-glycosite, whereas the glycan structure attached to the site is not being analyzed. Finally, releasing formerly N-linked glycopeptides from their attached oligosaccharide coalesces different oligosaccharide structures into a single peptide sequence-specific signal, thus obfuscating glycosylation changes owing to oligosaccharide structure alteration2.
SPEG has been successfully applied to the quantitative analysis of glycopeptides from complex mixtures, including serum/plasma and other body fluids5,6,11,12,13,14,15, the detection of glycoproteins secreted or shed into the culture medium by cells, the detection of membrane proteins and cell-surface proteins from cells and tissues16,17 and for comparing glycoproteins in the extracellular matrix of normal and disease tissues18. Several high-abundance plasma proteins including the most abundant plasma protein, albumin, do not appear to contain any N-glycosites and are therefore transparent to the method, allowing for more efficient identification and quantification of the lower abundance glycoproteins in blood6. By combining quantitative analysis of N-glycosites with methods that determine relative protein changes in different proteomes, such as the cysteine tagging method19, the occupancy of individual N-glycosites and changes therein can also be determined. This is of particular interest in studies in which changes of glycosylation occupancy are suspected, as exemplified by patients with type I congenital disorders of glycosylation, in which the N-linked glycosylation pathway is deficient20. Using this method, we have identified thousands of N-glycosites from different tissues, plasma and other body fluids, and established a database (http://www.unipep.org) as a public resource for glycoprotein analysis10.
Materials
Reagents
-
Hydrazide resin: Affi-Prep beads (Bio-Rad Laboratories)
-
Sodium periodate: make 100 mM in water before usage (21 mg in 1 ml of water)
Critical
This reagent is light sensitive.
-
Tris (2-carboxyethyl) phosphine: make 120 mM stock in water (Pierce, molecular weight=286.65, 34.4 mg in 1 ml of water)
-
Iodoacetamide: make 160 mM solution in water just before use (30 mg/ml in water, light sensitive)
-
Potassium phosphate buffer: 100 mM, pH 8.0
-
Ammonium bicarbonate (NH4HCO3): 0.1 M solution pH 8.3; make up fresh
-
Denaturing buffer: 8 M urea in 0.4 M NH4HCO3 with 0.1% (wt/vol) SDS
-
Sodium chloride: 1.5 M
-
SDS stock solution: 10% (wt/vol)
-
Standard peptide: 1 μM angiotensin I peptide in water (Sigma-Aldrich, cat. no. A9650) and 1 μM of neurotensin peptide in water (Sigma-Aldrich, cat. no. A6383)
-
Hydrochloric acid (HCl): 5 N
-
Acetic acid: 0.4% (vol/vol)
-
PNGase F (New England Biolabs)
-
Sequencing grade trypsin (Promega)
-
Acetonitrile (ACN)
-
BCA Protein Assay Kit (Pierce)
-
Methanol
-
SDS–PAGE gels
-
0.1% Trifluoroacetic acid (TFA) (vol/vol)
-
HPLC solution A: 0.1% (vol/vol) formic acid in water
-
HPLC solution B: 0.1% (vol/vol) formic acid in ACN
-
Succinic-d0-anhydride (Sigma)
-
Succinic-d4-anhydride: C/D/N Isotopes (Pointe-Claire)
-
Isotopic labeling solution: 2 mg ml−1 of succinic anhydride in dimethylformamide (DMF)/pyridine/H2O=50/10/40 (v/v/v)
-
Software for sequence assignment from tandem mass spectrum: SEQUEST21
-
Software for statistical evaluation of peptide sequence assignment: PeptideProphet22
Equipment
-
Tube rocker
-
SpeedVac
-
C18 1cc SepPak columns (Waters)
-
Vial: Glass vials with polyethylene snap cap (Waters)
-
HPLC: HP1100 (Agilent Technologies)
-
Sonicator
-
LCQ or LTQ ion-trap mass spectrometer (Thermo Finnigan)
-
Quadrupole-time-of-flight (electrospray ionization (ESI)-qTOF) mass spectrometer (Waters)
-
Peptide cartridge packed with Magic C18 (Michrom Bioresources)
-
FAMOS autosampler (DIONEX)
-
Microcapillary HPLC column: 10 cm × 75 μm i.d. packed with Magic C18 resin (5 μm, 100 Å; Michrom Bioresources)
Equipment Setup
-
LC-MS-MS/MS Peptides are injected into a peptide cartridge packed with Magic C18 using a FAMOS autosampler and then passed through a 10 cm × 75 μm i.d. microcapillary HPLC column packed with Magic C18 resin. The eluting peptides are directly ionized by ESI and tandem mass spectra (MS/MS) are acquired by data-dependent MS/MS mode (a full-scan mass spectrum is followed by a tandem mass spectrum), where the precursor ion is selected “on the fly” from the previous scan. A selected precursor is placed in a list and dynamically excluded for 3 min from further fragmentation.
-
HPLC A linear gradient of ACN from 5% (vol/vol) to 32% (vol/vol) over 100 min at a flow rate of ∼300 nl min−1 is applied for reverse-phase liquid chromatography using an HP1100 solvent delivery system.
-
HPLC gradient:
Table 1 Table 2
Procedure
Digestion of proteins to peptides
-
1
Extract proteins from a variety of biological samples in denaturing buffer (Fig. 2). For plasma proteins or other protein samples with a protein concentration of more than 5 mg ml−1, dilute the sample at least ten times with denaturing buffer (final protein concentration is less than 5 mg ml−1). For proteins from other body fluids or cell culture medium with protein concentration less than 5 mg ml−1, add solid urea, ammonium bicarbonate and 10% SDS stock solution directly to the samples to prepare a denaturing buffer containing 8 M urea, 0.4 M NH4HCO3 and 0.1% SDS. For extraction of protein from cells, harvest 107 cells in 1 ml of denaturing buffer. For extraction of proteins from solid tissue, frozen tissue (100 mg) is sliced into 1–3 mm3 thick pieces and incubated in 1 ml of urea buffer for 2–3 min with vortexing.
-
2
Sonicate samples for 6 min at 4 °C with a probe sonicator to homogenize protein samples.
-
3
Determine protein concentration with BCA protein assay, and take 1 mg of proteins from crude extract for each SPEG. More protein can be used for larger scale glycopeptide preparations if reagent amounts are scaled appropriately.
-
4
Add Tris (2-carboxyethyl) phosphine to a final concentration of 10 mM and incubate samples at 60 °C for 60 min.
-
5
Add iodoacetamide solution to a final concentration of 12 mM and incubate at room temperature (20 °C) in the dark for 30 min.
Critical Step
Denature proteins to ensure complete digestion of proteins to peptides.
-
6
Dilute proteins with phosphate buffer to reduce the urea concentration to less than 2 M and save ∼1 μg of protein to check on SDS–PAGE.
-
7
Add 20 μg of trypsin and digest samples at 37 °C for 4 h with gentle shaking.
Pause point
Digestion can be left overnight at 37 °C.
-
8
Remove undigested materials by centrifugation at 12,000g for 10 min.
-
9
Analyze ∼1 μg of peptides after digestion and the saved ∼1 μg of protein from Step 6 by SDS–PAGE to check the degree of completion of tryptic digestion.
Critical Step
Digest proteins completely for high specificity and yield of glycopeptide. If digestion is not complete, another batch of enzyme can be added, and digest samples for an additional 4 h at 37 °C.
Pause point
Digested peptides can be stored frozen at −20 °C for several weeks.
Coupling of glycopeptides to solid support
-
10
Add 10 μl of 5 N HCl to digested peptides (check pH ≤3).
-
11
Condition C18 SepPak column by washing C18 column twice with 1 ml of 0.1% TFA in 50% ACN and then twice with 1 ml 0.1% TFA.
-
12
Load the sample onto the conditioned C18 SepPak column.
-
13
Wash the C18 SepPak column three times with 1 ml of 0.1% TFA and elute the peptides twice with 0.2 ml of 0.1% TFA in 50% ACN.
-
14
Add 45 μl of sodium periodate to each sample. Incubate samples in dark at 4 °C for 1 h (10 mM final sodium periodate concentration).
-
15
Add 3.6 ml of 0.1% TFA to the sample.
-
16
Condition C18 SepPak column by washing C18 column twice with 1 ml of 0.1% TFA in 50% ACN and then twice with 1 ml 0.1% TFA.
-
17
Load the sample onto the conditioned C18 SepPak column.
-
18
Wash the C18 SepPak column three times with 0.1% TFA and elute the peptides twice with 0.2 ml of 80% ACN with 0.1% TFA.
-
19
Prepare 25 μl of pure hydrazide resin (50 μl of 50% slurry) per sample, spin the resin briefly (3,000 r.p.m. for 30 s) and remove the solution from the resin. Wash hydrazide resin by resuspending the resin in 1 ml of deionized water and removing water after brief spinning.
-
20
Add the oxidized peptides to the hydrazide resin and conjugate the glycopeptides at room temperature by mixing for a minimum of 3 h.
Pause point
Coupling can be left overnight at room temperature and immobilized glycopeptides on solid support can be stored at 4 °C for up to a month.
Labeling of N-linked glycopeptides with stable isotope tags
-
21
To detect quantitative changes of N-linked glycopeptides from two biological samples using isotopic labeling and mass spectrometry, wash the resin three times each with 800 μl of 1.5 M NaCl, water and DMF/pyridine/H2O=50/10/40 (v/v/v). Remove the supernatant by spinning at 2,500g for 5 min at each wash.
-
22
Resuspend the resin in 25 μl of DMF/pyridine/H2O=50/10/40 (v/v/v) and spike in 1 μl of a standard peptide with a free α-amino group (angiotensin I peptide) and 1 μl of standard peptide with a free ε-amino group in lysine (Neurotensin peptide).
-
23
Add light (d0) succinic anhydride solution to one biological sample and heavy (d4) isotope-labeled succinic anhydride solution to the second of the two samples being compared. The final concentration of succinic anhydride is 100 μM and the samples are incubated at room temperature (20 °C) for or 1 h.
Critical Step
Check the completion of peptide labeling by monitoring the mass shift of the standard peptide by mass spectrometry. Complete conversion of standard peptides to modified peptides (100 mass unit addition for labeling with light succinic anhydride and 104 mass unit addition for labeling with heavy succinic anhydride) is expected for complete labeling reactions. If necessary, incubate samples for another hour with an additional aliquot of succinic anhydride solution.
Releasing formerly N-glycosites from solid support
-
24
Wash the resin three times each with 800 μl of DMF, water and ammonium bicarbonate buffer. Remove the supernatant by spinning at 2,500g for 5 min at each step.
-
25
Resuspend the resin in 25 μl ammonium biocarbonate buffer, add 3 μl of PNGase F (500 U μl−1) and incubate at 37 °C for 4 h with mixing.
-
26
Transfer the supernatant to a glass vial and wash the hydrazide resin twice with 100 μl of ammonium bicarbonate buffer.
-
27
Combine the washes with the supernatant and purify the peptides with C18 SepPak column as described in Steps 10–13.
-
28
Dry the released glycopeptides in the glass vial within a SpeedVac in room temperature (20 °C) until complete dryness.
-
29
Dissolve peptides in 20 μl of 0.4% acetic acid for mass spectrometry analysis (5 μl can be used in each analysis).
Pause point
Released peptides can be stored at −20 °C.
Identification of peptides by mass spectrometry
-
30
Analyze peptides using LCQ or LTQ ion-trap mass spectrometer or ESI qTOF mass spectrometer according to the standard practices and manufacturers' instructions6.
-
31
Search MS/MS spectra against a protein database using the SEQUEST software21. The database search parameters are set to the following modifications: carboxymethylated cysteines, oxidized methionines, succinic anhydride modified amino-termini and lysines, and a (PNGase F-catalyzed) conversion of Asn to Asp that occurs at the original site of carbohydrate attachment to the peptide/protein (i.e., the N-glycosite). No other constraints are included for database searches.
-
32
Statistically analyze database search results using PeptideProphet, which effectively computes a probability for the likelihood of each identification being correct (on a scale of 0–1) in a data-dependent fashion22. A PeptideProphet probability score of ≥0.9 is used as a filter to remove low-probability peptide identifications. As it is known that the majority of N-linked glycosylation occurs at a consensus N-X-S/T sequon (where X is any amino acid except proline)9, the assigned peptide sequences from Step 31 are additionally filtered to remove non-motif-containing peptides. Finally, peptide sequences are analyzed with respect to individual unique N-X-S/T sequons such that overlapping sequences containing the same N-X-S/T sequon (i.e., redundant N-linked glycopeptides for the same N-glycosite) are resolved in favor of those peptide sequences that contained the greater number of tryptic cleavage termini.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Timing
Steps 1–9: up to 1 day
Steps 10–20: up to 1 day
Steps 21–29: up to 1 day
Steps 30–32: up to 1 day
Anticipated results
To illustrate the protocol, glycopeptides from 1 mg of proteins from tissues and plasma were isolated, and ∼10 μg of total N-linked glycopeptides was recovered. Such isolated peptides were analyzed by LTQ and MS/MS spectra were searched against protein database using SEQUEST. The analysis resulted in over 1,000 peptide identifications that contain the consensus N-linked glycosylation motif (N-X-S/T motif, X can be any amino acid except proline) and have a PeptideProphet score of at least 0.9 (ref. 6). The identified glycopeptides represent over 300 individual, unique N-X-S/T sequons after overlapping sequences containing the same N-X-S/T sequon are resolved. The specificity of the identified peptides is over 90%, as calculated by the percentage of identified peptides containing consensus N-linked glycosylation motif.
Among about 300 peptides identified from the mixed samples, 28 peptides were detected and quantified in both tissue and plasma. These results indicate that the method can selectively isolate N-glycosites from mixtures of glycoproteins.
An example of quantitative analysis of glycopeptides with isotopic labeling is shown in Figure 3 (ref. 6). Glycopeptides immobilized on hydrazide resin from two samples were labeled with light (d0) or heavy (d4) forms of succinic anhydride. After labeling, the beads containing the two samples were combined and the formerly N-glycopeptides were released. The combined samples were analyzed by LC-MS using ESI-qTOF. The quantification is illustrated for a single scan of the mass spectrometer in MS mode. The paired peptide is doublely charged and has a mass difference of 4 U with monoisotopic peaks at 629.88 and 631.86 (Fig. 3). The peptide was sequenced by MS/MS analysis using ESI-qTOF and identified by database searching21. It was identified with peptide sequence EEQFN#STFR from human Ig γ-1 chain C region secreted form, a classic serum protein. This shows that accurate quantification of relative glycopeptide abundance from two samples can be achieved with stable isotope tagging.
References
Spiro, R.G. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R (2002).
Durand, G. & Seta, N. Protein glycosylation and diseases: blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin. Chem. 46, 795–805 (2000).
Freeze, H.H. Update and perspectives on congenital disorders of glycosylation. Glycobiology 11, 129R–143R (2001).
Kaji, H. et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 21, 667–672 (2003).
Zhang, H., Li, X.J., Martin, D.B. & Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 (2003).
Zhang, H. et al. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol. Cell. Proteomics 4, 144–155 (2005).
Anderson, N.L. & Anderson, N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 (2002).
Roth, J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem. Rev. 102, 285–303 (2002).
Bause, E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem. J. 209, 331–336 (1983).
Zhang, H. et al. UniPep, a database for human N-linked glycosites: a resource for biomarker discovery. Genome Biol. 7, R73 (2006).
Deutsch, E.W. et al. Human Plasma PeptideAtlas. Proteomics 5, 3497–3500 (2005).
Li, X.J. et al. A software suite for the generation and comparison of Peptide arrays from sets of data collected by liquid chromatography–mass spectrometry. Mol. Cell. Proteomics 4, 1328–1340 (2005).
Liu, T. et al. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 4, 2070–2080 (2005).
Ramachandran, P. et al. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 5, 1493–1503 (2006).
Pan, S. et al. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J. Proteome Res. 5, 2769–2779 (2006).
Zhang, H. et al. Mass spectrometric detection of tissue proteins in plasma. Mol. Cell. Proteomics. 6, 64–71 (2007).
Lewandrowski, U., Moebius, J., Walter, U. & Sickmann, A. Elucidation of N-glycosylation sites on human platelet proteins: a glycoproteomic approach. Mol. Cell. Proteomics 5, 226–233 (2006).
Liu, A.Y., Zhang, H., Sorensen, C.M. & Diamond, D.L. Analysis of prostate cancer by proteomics using tissue specimens. J. Urol. 173, 73–78 (2005).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Aebi, M. & Hennet, T. Congenital disorders of glycosylation: genetic model systems lead the way. Trends Cell Biol. 11, 136–141 (2001).
Eng, J., McCormack, A.L. & Yates, J.R., III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Acknowledgements
This work was supported with federal funds from the National Cancer Institute, National Institutes of Health, by grant R21-CA-114852 (to H.Z.) and by Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract no. N01-HV-8179 (to R. A.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Tian, Y., Zhou, Y., Elliott, S. et al. Solid-phase extraction of N-linked glycopeptides. Nat Protoc 2, 334–339 (2007). https://doi.org/10.1038/nprot.2007.42
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.42
This article is cited by
-
Identification of potential molecular targets for the treatment of cluster 1 human pheochromocytoma and paraganglioma via comprehensive proteomic characterization
Clinical Proteomics (2023)
-
A novel graphene oxide/chitosan foam incorporated with metal–organic framework stationary phase for simultaneous enrichment of glycopeptide and phosphopeptide with high efficiency
Analytical and Bioanalytical Chemistry (2022)
-
Proteomics of protein trafficking by in vivo tissue-specific labeling
Nature Communications (2021)
-
Discovery proteomics defines androgen-regulated glycoprotein networks in prostate cancer cells, as well as putative biomarkers of prostatic diseases
Scientific Reports (2021)
-
N-linked glycosylation enzymes in the diatom Thalassiosira oceanica exhibit a diel cycle in transcript abundance and favor for NXT-type sites
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.