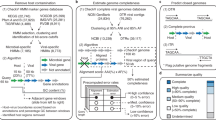
Abstract
The analysis of large microbiome data sets holds great promise for the delineation of the biological and metabolic functioning of living organisms and their role in the environment. In the midst of this genomic puzzle, viruses, especially those that infect microbial communities, represent a major reservoir of genetic diversity with great impact on biogeochemical cycles and organismal health. Overcoming the limitations associated with virus detection directly from microbiomes can provide key insights into how ecosystem dynamics are modulated. Here, we present a computational protocol for accurate detection and grouping of viral sequences from microbiome samples. Our approach relies on an expanded and curated set of viral protein families used as bait to identify viral sequences directly from metagenomic assemblies. This protocol describes how to use the viral protein families catalog (∼7 h) and recommended filters for the detection of viral contigs in metagenomic samples (∼6 h), and it describes the specific parameters for a nucleotide-sequence-identity-based method of organizing the viral sequences into quasi-species taxonomic-level groups (∼10 min).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Chen, I.A. et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516 (2017).
Mukherjee, S. et al. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 45, D446–D456 (2017).
Angly, F.E. et al. The marine viromes of four oceanic regions. PLoS Biol. 4, e368 (2006).
Breitbart, M., Miyake, J.H. & Rohwer, F. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236, 249–256 (2004).
Breitbart, M. & Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284 (2005).
Marhaver, K.L., Edwards, R.A. & Rohwer, F. Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10, 2277–2286 (2008).
Suttle, C.A., Chan, A.M. & Cottrell, M.T. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton 57, 721–726 (1991).
Dell'Anno, A., Corinaldesi, C., Magagnini, M. & Danovaro, R. Determination of viral production in aquatic sediments using the dilution-based approach. Nat. Protoc. 4, 1013–1022 (2009).
Thurber, R.V., Haynes, M., Breitbart, M., Wegley, L. & Rohwer, F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4, 470–483 (2009).
Brum, J.R. et al. Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015).
Dinsdale, E.A. et al. Functional metagenomic profiling of nine biomes. Nature 452, 629–632 (2008).
Mizuno, C.M., Rodriguez-Valera, F., Kimes, N.E. & Ghai, R. Expanding the marine virosphere using metagenomics. PLoS Genet. 9, e1003987 (2013).
Roux, S. et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693 (2016).
Akhter, S., Aziz, R.K. & Edwards, R.A. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 40, e126 (2012).
Fouts, D.E. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 34, 5839–5851 (2006).
Lima-Mendez, G., Van Helden, J., Toussaint, A. & Leplae, R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24, 863–865 (2008).
Arndt, D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016).
Roux, S., Enault, F., Hurwitz, B.L. & Sullivan, M.B. VirSorter: mining viral signal from microbial genomic data. PeerJ 3, e985 (2015).
Grazziotin, A.L., Koonin, E.V. & Kristensen, D.M. Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation. Nucleic Acids Res. 45, D491–D498 (2017).
Paez-Espino, D. et al. Uncovering earth's virome. Nature 536, 425–430 (2016).
Ivanova, N. et al. A call for standardized classification of metagenome projects. Environ. Microbiol. 12, 1803–1805 (2010).
Mukherjee, S. et al. Genomes OnLine Database(GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 45, D446–D456 (2016).
Paez-Espino, D. et al. IMG/VR: a database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res. 45, D457–D465 (2017).
Merchant, N. et al. The iPlant Collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 14, e1002342 (2016).
Suttle, C.A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Edwards, R.A., McNair, K., Faust, K., Raes, J. & Dutilh, B.E. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol. Rev. 40, 258–272 (2016).
Villarroel, J. et al. HostPhinder: a phage host prediction tool. Viruses 8 http://dx.doi.org/10.3390/v8050116 (2016).
Goren, M.G., Yosef, I. & Qimron, U. Programming bacteriophages by swapping their specificity determinants. Trends Microbiol. 23, 744–746 (2015).
Salmond, G.P. & Fineran, P.C. A century of the phage: past, present and future. Nat. Rev. Microbiol. 13, 777–786 (2015).
Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Enright, A.J., Van Dongen, S. & Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Finn, R.D., Clements, J. & Eddy, S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Chen, I.A. et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516 (2016).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Dutilh, B.E. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498 (2014).
Aziz, R.K., Dwivedi, B., Akhter, S., Breitbart, M. & Edwards, R.A. Multidimensional metrics for estimating phage abundance, distribution, gene density, and sequence coverage in metagenomes. Front. Microbiol. 6, 381 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Langdon, W.B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 8, 1 (2015).
Finn, R.D. et al. HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 (2015).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Li, D., Liu, C.M., Luo, R., Sadakane, K. & Lam, T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Dick, G.J. et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 10, R85 (2009).
Oulas, A. et al. Metagenomic investigation of the geologically unique Hellenic volcanic arc reveals a distinctive ecosystem with unexpected physiology. Environ. Microbiol. 18, 1122–1136 (2016).
Price, M.N., Dehal, P.S. & Arkin, A.P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009).
Huson, D.H. & Scornavacca, C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 (2012).
Acknowledgements
This work was supported by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, under contract no. DE-AC02-05CH11231, and used resources of the National Energy Research Scientific Computing Center, supported by the Office of Science of the US Department of Energy.
Author information
Authors and Affiliations
Contributions
D.P.-E., N.N.I., and N.C.K. conceived and led the protocol. G.A.P. provided computational and scripting support. All authors wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Details of the protocol for the given example.
Pipeline of the workflow including the name of all the files generated during the virus detection for the sampleidentified as 3300001348 in IMG/M (in blue), as well as the approximate time necessary for each of the stepsof the protocol (in red), and required scripts (bold black). The three yellow boxes indicate the three final outputsof this exercise: (i) 640 unique metagenomic viral contigs (mVCs) detected; (ii) 246 viral groups that include268 mVCs (out of the 640) from the given example as well as 457 metagenomic viral contigs from 32 otherdifferent metagenomes, and (iii) a list of 12,963 viral sequences of low abundance (from 8,436 unique viralgroups) with at least 10% of their length covered by unassembled reads (>90% sequence identity) from the targeted metagenome.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 and Supplementary Tables 1 and 2. (PDF 13452 kb)
Rights and permissions
About this article
Cite this article
Paez-Espino, D., Pavlopoulos, G., Ivanova, N. et al. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat Protoc 12, 1673–1682 (2017). https://doi.org/10.1038/nprot.2017.063
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.063
This article is cited by
-
The gut ileal mucosal virome is disturbed in patients with Crohn’s disease and exacerbates intestinal inflammation in mice
Nature Communications (2024)
-
Hidden diversity and potential ecological function of phosphorus acquisition genes in widespread terrestrial bacteriophages
Nature Communications (2024)
-
Altered human gut virome in patients undergoing antibiotics therapy for Helicobacter pylori
Nature Communications (2023)
-
Soil viral diversity, ecology and climate change
Nature Reviews Microbiology (2023)
-
Genomic diversity and ecological distribution of marine Pseudoalteromonas phages
Marine Life Science & Technology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.