Abstract
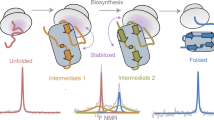
During biosynthesis on the ribosome, an elongating nascent polypeptide chain can begin to fold, in a process that is central to all living systems. Detailed structural studies of co-translational protein folding are now beginning to emerge; such studies were previously limited, at least in part, by the inherently dynamic nature of emerging nascent chains, which precluded most structural techniques. NMR spectroscopy is able to provide atomic-resolution information for ribosome–nascent chain complexes (RNCs), but it requires large quantities (≥10 mg) of homogeneous, isotopically labeled RNCs. Further challenges include limited sample working concentration and stability of the RNC sample (which contribute to weak NMR signals) and resonance broadening caused by attachment to the large (2.4-MDa) ribosomal complex. Here, we present a strategy to generate isotopically labeled RNCs in Escherichia coli that are suitable for NMR studies. Uniform translational arrest of the nascent chains is achieved using a stalling motif, and isotopically labeled RNCs are produced at high yield using high-cell-density E. coli growth conditions. Homogeneous RNCs are isolated by combining metal affinity chromatography (to isolate ribosome-bound species) with sucrose density centrifugation (to recover intact 70S monosomes). Sensitivity-optimized NMR spectroscopy is then applied to the RNCs, combined with a suite of parallel NMR and biochemical analyses to cross-validate their integrity, including RNC-optimized NMR diffusion measurements to report on ribosome attachment in situ. Comparative NMR studies of RNCs with the analogous isolated proteins permit a high-resolution description of the structure and dynamics of a nascent chain during its progressive biosynthesis on the ribosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cabrita, L.D. et al. A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat. Struct. Mol. Biol. 23, 278–285 (2016).
Cabrita, L.D., Dobson, C.M. & Christodoulou, J. Protein folding on the ribosome. Curr. Opin. Struct. Biol. 20, 33–45 (2010).
Nicola, A.V., Chen, W. & Helenius, A. Co-translational folding of an alphavirus capsid protein in the cytosol of living cells. Nat. Cell Biol. 1, 341–345 (1999).
Hoffmann, A. et al. Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol. Cell 48, 63–74 (2012).
Ito, K. & Chiba, S. Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 82, 171–202 (2013).
Kelkar, D.A., Khushoo, A., Yang, Z. & Skach, W.R. Kinetic analysis of ribosome-bound fluorescent proteins reveals an early, stable, cotranslational folding intermediate. J. Biol. Chem. 287, 2568–2578 (2012).
Kim, S.J. et al. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 348, 444–448 (2015).
Seidelt, B. et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326, 1412–1415 (2009).
Bhushan, S. et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 9, e1000581 (2011).
Cabrita, L.D., Hsu, S.-T.D., Launay, H., Dobson, C.M. & Christodoulou, J. Probing ribosome-nascent chain complexes produced in vivo by NMR spectroscopy. Proc. Natl. Acad. Sci. USA 106, 22239–22244 (2009).
Evans, M.S., Ugrinov, K.G., Frese, M.-A. & Clark, P.L. Homogeneous stalled ribosome nascent chain complexes produced in vivo or in vitro. Nat. Methods 2, 757–762 (2005).
Nakatogawa, H. & Ito, K. The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636 (2002).
Ugrinov, K.G. & Clark, P.L. Cotranslational folding increases GFP folding yield. Biophys. J. 98, 1312–1320 (2010).
Kudlicki, W., Chirgwin, J., Kramer, G. & Hardesty, B. Folding of an enzyme into an active conformation while bound as peptidyl-tRNA to the ribosome. Biochemistry 34, 14284–14287 (1995).
Evans, M.S., Sander, I.M. & Clark, P.L. Cotranslational folding promotes beta-helix formation and avoids aggregation in vivo. J. Mol. Biol. 383, 683–692 (2008).
Tsalkova, T., Odom, O.W., Kramer, G. & Hardesty, B. Different conformations of nascent peptides on ribosomes. J. Mol. Biol. 278, 713–723 (1998).
Kleizen, B., van Vlijmen, T., de Jonge, H.R. & Braakman, I. Folding of CFTR is predominantly cotranslational. Mol. Cell 20, 277–287 (2005).
Frydman, J., Erdjument-Bromage, H., Tempst, P. & Hartl, F.U. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat. Struct. Biol. 6, 697–705 (1999).
Friguet, B., Djavadi-Ohaniance, L., King, J. & Goldberg, M.E. In vitro and ribosome-bound folding intermediates of P22 tailspike protein detected with monoclonal antibodies. J. Biol. Chem. 269, 15945–15949 (1994).
Speed, M.A., Morshead, T., Wang, D.I. & King, J. Conformation of P22 tailspike folding and aggregation intermediates probed by monoclonal antibodies. Protein Sci. 6, 99–108 (1997).
Zhang, G., Hubalewska, M. & Ignatova, Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274–280 (2009).
Holtkamp, W. et al. Cotranslational protein folding on the ribosome monitored in real time. Science 350, 1104–1107 (2015).
Woolhead, C.A., McCormick, P.J. & Johnson, A.E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 (2004).
Lu, J. & Deutsch, C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry 44, 8230–8243 (2005).
Kaiser, C.M., Goldman, D.H., Chodera, J.D., Tinoco, I. & Bustamante, C. The ribosome modulates nascent protein folding. Science 334, 1723–1727 (2011).
Knight, A.M. et al. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem. Biol. 8, 1195–1204 (2013).
Sander, I.M., Chaney, J.L. & Clark, P.L. Expanding Anfinsen's principle: contributions of synonymous codon selection to rational protein design. J. Am. Chem. Soc. 136, 858–861 (2014).
Fedyunin, I. et al. tRNA concentration fine tunes protein solubility. FEBS Lett. 586, 3336–3340 (2012).
Corazza, A. et al. Native-unlike long-lived intermediates along the folding pathway of the amyloidogenic protein beta2-microglobulin revealed by real-time two-dimensional NMR. J. Biol. Chem. 285, 5827–5835 (2010).
Kaiser, C.M. et al. Real-time observation of trigger factor function on translating ribosomes. Nature 444, 455–460 (2006).
Nilsson, O.B., Müller-Lucks, A., Kramer, G., Bukau, B. & von Heijne, G. Trigger factor reduces the force exerted on the nascent chain by a cotranslationally folding protein. J. Mol. Biol. 428, 1356–1364 (2016).
Gabashvili, I.S. et al. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell 100, 537–549 (2000).
Stark, H. et al. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389, 403–406 (1997).
Nissen, P., Hansen, J., Ban, N., Moore, P.B. & Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000).
Ban, N., Nissen, P., Hansen, J., Moore, P.B. & Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Gao, Y.-G. et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Bhushan, S. et al. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 17, 313–317 (2010).
Hsu, S.-T.D. et al. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc. Natl. Acad. Sci. USA 104, 16516–16521 (2007).
Hsu, S.T., Cabrita, L.D., Fucini, P., Christodoulou, J. & Dobson, C.M. Probing side-chain dynamics of a ribosome-bound nascent chain using methyl NMR spectroscopy. J. Am. Chem. Soc. 131, 8366–8367 (2009).
Eichmann, C., Preissler, S., Riek, R. & Deuerling, E. Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Proc. Natl. Acad. Sci USA 107, 9111–9116 (2010).
Hsu, S.T.D. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc. Natl. Acad. Sci. USA 104, 16516–16521 (2007).
Nilsson, O.B. et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 12, 1533–1540 (2015).
Christodoulou, J. et al. Heteronuclear NMR investigations of dynamic regions of intact Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA 101, 10949–10954 (2004).
Mulder, F.A. et al. Conformation and dynamics of ribosomal stalk protein L12 in solution and on the ribosome. Biochemistry 43, 5930–5936 (2004).
Rutkowska, A. et al. Large-scale purification of ribosome-nascent chain complexes for biochemical and structural studies. FEBS Lett. 583, 2407–2413 (2009).
Hsu, S.T., Cabrita, L.D., Fucini, P., Dobson, C.M. & Christodoulou, J. Structure, dynamics and folding of an immunoglobulin domain of the gelation factor (ABP-120) from Dictyostelium discoideum. J. Mol. Biol. 388, 865–879 (2009).
Deckert, A.W. et al. Structural characterization of the interaction of α-synuclein nascent chains with the ribosomal surface and trigger factor. Proc. Natl. Acad. Sci. USA 113, 5012–5017 (2016).
Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Spedding, G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. Ribosomes and Protein Synthesis. 1–29 (IRL Press, 1990).
Bergemann, K. & Nierhaus, K.H. Spontaneous, elongation factor G independent translocation of Escherichia coli ribosomes. J. Biol. Chem. 258, 15105–15113 (1983).
Fredrick, K. & Noller, H.F. Catalysis of ribosomal translocation by sparsomycin. Science 300, 1159–1162 (2003).
Bai, Y., Milne, J.S., Mayne, L. & Englander, S.W. Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 (1993).
Deusser, E. & Wittmann, H.G. Ribosomal proteins: variation of the protein composition in Escherichia coli ribosomes as function of growth rate. Nature 238, 269–270 (1972).
Rostom, A.A. et al. Detection and selective dissociation of intact ribosomes in a mass spectrometer. Proc. Natl. Acad. Sci. USA 97, 5185–5190 (2000).
Ramagopal, S. & Subramanian, A.R. Alteration in the acetylation level of ribosomal protein L12 during growth cycle of Escherichia coli. Proc. Natl. Acad. Sci. USA 71, 2136–2140 (1974).
Lu, J. & Deutsch, C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 12, 1123–1129 (2005).
Chan, S.H.S., Waudby, C.A., Cassaignau, A.M.E., Cabrita, L.D. & Christodoulou, J. Increasing the sensitivity of NMR diffusion measurements by paramagnetic longitudinal relaxation enhancement, with application to ribosome-nascent chain complexes. J. Biomol. NMR 63, 1–13 (2015).
Kay, L.E. Solution NMR spectroscopy of supra-molecular systems, why bother? A methyl-TROSY view. J. Magn. Reson. 210, 159–170 (2011).
Wiesner, S. & Sprangers, R. Methyl groups as NMR probes for biomolecular interactions. Curr. Opin. Struct. Biol. 35, 60–67 (2015).
Tugarinov, V., Kanelis, V. & Kay, L.E. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 1, 749–754 (2006).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Vranken, W.F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005).
Sivashanmugam, A. et al. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 18, 936–948 (2009).
Hill, W.E., Rossetti, G.P. & Van Holde, K.E. Physical studies of ribosomes from Escherichia coli. J. Mol. Biol. 44, 263–277 (1969).
Mahmood, T. & Yang, P.-C. Western blot: technique, theory, and trouble shooting. N. Am. J. Med. Sci. 4, 429–434 (2012).
Stejskal, E.O. & Tanner, J.E. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J. Chem. Phys. 42, 288 (1965).
Wu, D.H., Chen, A.D. & Johnson, C.S. An improved diffusion-ordered spectroscopy experiment incorporating bipolar-gradient pulses. J. Magn. Reson. A 115, 260–264 (1995).
Augustyniak, R., Ferrage, F., Damblon, C., Bodenhausen, G. & Pelupessy, P. Efficient determination of diffusion coefficients by monitoring transport during recovery delays in NMR. Chem. Commun. 48, 5307–5309 (2012).
Waudby, C.A., Launay, H., Cabrita, L.D. & Christodoulou, J. Protein folding on the ribosome studied using NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 74, 57–75 (2013).
Waudby, C.A. & Christodoulou, J. An analysis of NMR sensitivity enhancements obtained using non-uniform weighted sampling, and the application to protein NMR. J. Magn. Reson. 219, 46–52 (2012).
Acknowledgements
We thank Bernd Bukau (Ruprecht-Karls-Universität Heidelberg) for anti-SecM antibodies. We acknowledge the use of the ISMB Biological NMR Facility, UCL. We thank T. Frenkiel, G. Kelly and A. Oregioni and acknowledge the use of the biomolecular NMR facilities of the MRC NMR Centre at the Francis Crick Institute. This work was supported by a New Investigator Award (BBSRC BBG0156511, to J.C.), a Wellcome Trust Investigator Award (097806/Z/11/Z, to J.C.) and an AlphaOne Foundation grant (to L.D.C.); A.L.R. is an NHMRC CJ Martin Fellow.
Author information
Authors and Affiliations
Contributions
A.M.E.C., H.M.M.L., C.A.W., J.C. and L.D.C. designed the study. A.M.E.C., H.M.M.L., M.-E.K., X.W., C.A.W., A.D., A.L.R. and L.D.C. performed the research. A.M.E.C., C.A.W., H.M.M.L., J.C. and L.D.C. wrote the paper. All authors discussed the results and contributed to the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 2D correlation spectra of FLN5 in the presence of 70S ribosomes
a SOFAST 2D correlation spectrum overlay of 5 μM 15N-labelled FLN5 in the absence (red) and presence (cyan) of 5 μM 70S ribosomes. b The relative intensities of an equimolar sample of FLN5 and 70S ribosomes (5 µM) vs. FLN5 alone (5 µM), plotted against primary sequence, shows that the intensities of FLN5 resonances remain unchanged in the presence of ribosomes, making it a tractable system for RNC studies. The analogous study can be undertaken with the unfolded polypeptide. All uncertainties have been derived from the spectral noise. Adapted from ref. 1 with permission from Nature Publishing Group.
Supplementary Figure 2 Assessment of ribosome background labelling
To assess the level of ribosomal background labelling within RNC samples, 15N/13C filtered 1H 1D (red) and 15N/13C edited 1H 1D (grey) spectra are collected. The intensity of the 1H envelope of bL12 resonances bound to 14N (15N/13C filtered 1H 1D) is matched by scaling to that of 15N-bound protons (15N/13C edited 1H 1D) in order to quantify the ratio of unlabelled to labelled species. a Intensity profiles of amide resonances in 15N filtered 1H 1D (red) and 15N 1H edited 1D (grey) spectra from a 15N-labelled RNC, in which the background labelling amounts to ~6%. b Intensity profiles of methyl resonances in 13C filtered 1H 1D (red) and 13C edited 1H 1D (grey) spectra from a 13C-labelled RNC. We determine that ~13% of its bL12 resonances are labelled.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 392 kb)
Rights and permissions
About this article
Cite this article
Cassaignau, A., Launay, H., Karyadi, ME. et al. A strategy for co-translational folding studies of ribosome-bound nascent chain complexes using NMR spectroscopy. Nat Protoc 11, 1492–1507 (2016). https://doi.org/10.1038/nprot.2016.101
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.101
This article is cited by
-
Insights into the ribosome function from the structures of non-arrested ribosome–nascent chain complexes
Nature Chemistry (2023)
-
The ribosome stabilizes partially folded intermediates of a nascent multi-domain protein
Nature Chemistry (2022)
-
Modulating co-translational protein folding by rational design and ribosome engineering
Nature Communications (2022)
-
Nascent chains can form co-translational folding intermediates that promote post-translational folding outcomes in a disease-causing protein
Nature Communications (2021)
-
Interactions between nascent proteins and the ribosome surface inhibit co-translational folding
Nature Chemistry (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.