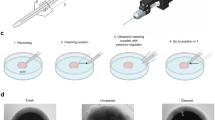
Abstract
Whole-cell patch clamping in vivo is an important neuroscience technique that uniquely provides access to both suprathreshold spiking and subthreshold synaptic events of single neurons in the brain. This article describes how to set up and use the autopatcher, which is a robot for automatically obtaining high-yield and high-quality whole-cell patch clamp recordings in vivo. By following this protocol, a functional experimental rig for automated whole-cell patch clamping can be set up in 1 week. High-quality surgical preparation of mice takes ∼1 h, and each autopatching experiment can be carried out over periods lasting several hours. Autopatching should enable in vivo intracellular investigations to be accessible by a substantial number of neuroscience laboratories, and it enables labs that are already doing in vivo patch clamping to scale up their efforts by reducing training time for new lab members and increasing experimental durations by handling mentally intensive tasks automatically.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Bruno, R.M. & Sakmann, B. Cortex is driven by weak but synchronously active thalamocortical synapses. Science 312, 1622–1627 (2006).
Arenz, A., Silver, R.A., Schaefer, A.T. & Margrie, T.W. The contribution of single synapses to sensory representation in vivo. Science 321, 977–980 (2008).
Brecht, M., Schneider, M., Sakmann, B. & Margrie, T.W. Whisker movements evoked by stimulation of single pyramidal cells in rat motor cortex. Nature 427, 704–710 (2004).
Chadderton, P., Margrie, T.W. & Hausser, M. Integration of quanta in cerebellar granule cells during sensory processing. Nature 428, 856–860 (2004).
Eberwine, J. et al. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA 89, 3010–3014 (1992).
Rancz, E.A. et al. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat. Neurosci. 14, 527–532 (2011).
Margrie, T.W., Brecht, M. & Sakmann, B. In vivo, low-resistance, whole-cell recordings from neurons in the anaesthetized and awake mammalian brain. Pflugers Arch. 444, 491–498 (2002).
Chadderton, P., Agapiou, J.P., McAlpine, D. & Margrie, T.W. The synaptic representation of sound source location in auditory cortex. J. Neurosci. 29, 14127–14135 (2009).
Chadderton, P., Margrie, T.W., Hausser, M Integration of quanta in cerebellar granule cells during sensory processing. Nature 428, 856–860 (2004).
Crochet, S. & Petersen, C.C.H. Correlating whisker behavior with membrane potential in barrel cortex of awake mice. Nat. Neurosci. 9, 608–610 (2006).
Crochet, S., Poulet, J.F., Kremer, Y. & Petersen, C.C. Synaptic mechanisms underlying sparse coding of active touch. Neuron 69, 1160–1175 (2011).
Gentet, L.J., Avermann, M., Matyas, F., Staiger, J.F. & Petersen, C.C. Membrane potential dynamics of GABAergic neurons in the barrel cortex of behaving mice. Neuron 65, 422–435 (2010).
Gentet, L.J. et al. Unique functional properties of somatostatin-expressing GABAergic neurons in mouse barrel cortex. Nat. Neurosci. 15, 607–612 (2012).
Harvey, C.D., Collman, F., Dombeck, D.A. & Tank, D.W. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature 461, 941–946 (2009).
Hromadka, T., DeWeese, M.R. & Zador, A.M. Sparse representation of sounds in the unanesthetized auditory cortex. PLoS Biol. 6, 124–137 (2008).
Schaefer, A.T. & Margrie, T.W. Spatiotemporal representations in the olfactory system. Trends Neurosci. 30, 92–100 (2007).
Kitamura, K., Judkewitz, B., Kano, M., Denk, W. & Hausser, M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat. Methods 5, 61–67 (2008).
Komai, S., Denk, W., Osten, P., Brecht, M. & Margrie, T.W. Two-photon targeted patching (TPTP) in vivo. Nat. Protoc. 1, 647–652 (2006).
Margrie, T.W. et al. Targeted whole-cell recordings in the mammalian brain in vivo. Neuron 39, 911–918 (2003).
Lee, A.K., Epsztein, J. & Brecht, M. Head-anchored whole-cell recordings in freely moving rats. Nat. Protoc. 4, 385–392 (2009).
Lee, A.K., Manns, I.D., Sakmann, B. & Brecht, M. Whole-cell recordings in freely moving rats. Neuron 51, 399–407 (2006).
Lee, D., Lin, B.-J. & Lee, A.K. Hippocampal place fields emerge upon single-cell manipulation of excitability during behavior. Science 337, 849–853 (2012).
Long, M.A., Jin, D.Z. & Fee, M.S. Support for a synaptic chain model of neuronal sequence generation. Nature 468, 394–399 (2010).
Kodandaramaiah, S.B., Franzesi, G.T., Chow, B.Y., Boyden, E.S. & Forest, C.R. Automated whole-cell patch-clamp electrophysiology of neurons in vivo. Nat. Methods 9, 585–587 (2012).
DeWeese, M.R. Whole-cell recording in vivo. Curr. Protoc. Neurosci. 38, 6.22.1–6.22.15 (2007).
Chuong, A.S. et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci. 17, 1123–1129 (2014).
Schramm, A.E., Marinazzo, D., Gener, T. & Graham, L.J. The touch and zap method for in vivo whole-cell patch recording of intrinsic and visual responses of cortical neurons and glial cells. PLoS ONE 9, e97310 (2014).
Pak, N. et al. Closed-loop, ultraprecise, automated craniotomies. J. Neurophysiol. 113, 3943–3953 (2015).
Kodandaramaiah, S.B., Boyden, E.S. & Forest, C.R. In vivo robotics: the automation of neuroscience and other intact-system biological fields. Ann. NY Acad. Sci. 1305, 63–71 (2013).
Arenkiel, B.R. et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron 54, 205–218 (2007).
Harrison, R.R. et al. Microchip amplifier for in vitro, in vivo, and automated whole-cell patch-clamp recording. J. Neurophysiol. 10.1152/jn.00629.2014 (2014).
Poulet, J.F.A., Fernandez, L.M.J., Crochet, S. & Petersen, C.C.H. Thalamic control of cortical states. Nat. Neurosci. 15, 370–372 (2012).
Polack, P.O., Friedman, J. & Golshani, P. Cellular mechanisms of brain state-dependent gain modulation in visual cortex. Nat. Neurosci. 16, 1331–1339 (2013).
Plant, T.D., Eilers, J. & Konnerth, A. in Patch-Clamp Applications and Protocols. Vol. 26 (eds. Boulton, A., Baker, G. & Walz, W.) 233–258 (Humana Press, 1995).
Acknowledgements
We thank B.D. Allen and H.-J. Suk for feedback on the manuscript. C.R.F. acknowledges the National Institutes of Health (NIH) BRAIN Initiative (National Eye Institute (NEI) and National Institute of Mental Health (NIMH) 1-U01-MH106027-01), an NIH Single Cell Grant 1 R01 EY023173, the National Science Foundation (NSF) (Education and Human Resources (Her) 0965945 and Computer and Information Science and Engineering (CISE) 1110947), an NIH Computational Neuroscience Training grant (no. 5T90DA032466), the Georgia Tech Translational Research Institute for Biomedical Engineering & Science (TRIBES) Seed Grant Awards Program, the Georgia Tech Fund for Innovation in Research and Education (GT-FIRE), the Wallace H. Coulter Translational/Clinical Research Grant Program and support from Georgia Tech through the Institute for Bioengineering and Biosciences Junior Faculty Award, the Technology Fee Fund, Invention Studio, and the George W. Woodruff School of Mechanical Engineering. E.S.B. acknowledges NIH 1R01EY023173, the New York Stem Cell Foundation-Robertson Award, a NIH Director's Pioneer Award 1DP1NS087724, an NIH Director's Transformative Award (NIH 1R01MH103910) and an NIH BRAIN initiative grant (NIH 1R24MH106075). G.T.F. acknowledges a Friends of the McGovern Institute Fellowship.
Author information
Authors and Affiliations
Contributions
S.B.K., I.R.W., G.L.H., C.R.F. and E.S.B. designed, built and tested the autopatcher system. A.C.S. and G.T.F. assisted with experiments. M.L.M. developed the software included with the manuscript. S.B.K., I.R.W., G.L.H., A.C.S., G.T.F., C.R.F. and E.S.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.C.S., G.T.F., M.L.M., C.R.F. and E.S.B. declare no competing interests. I.R.W., S.B.K. and G.L.H. received financial remuneration from Neuromatic Devices for technical consulting services provided in 2012, 2013 and 2012–2015, respectively.
Supplementary information
Supplementary Text and Figures
Supplementary Methods (PDF 176 kb)
Supplementary Data 1: File archive consisting of software required for running the Autopatcher.
Includes two Labview library files – ‘Autopatcher 2000.llb’ and ‘Hardware.llb’ that can be opened using Labview installed in Step 8 of the protocol. Also included is a corresponding ‘Autopatcher software configuration manual.pdf’ that provides detailed instructions on installation of software and configuring the software settings to control the autopatcher control box. (ZIP 2066 kb)
Supplementary Data 2: File archive consisting of mechanical drawings and computer aided design (CAD) files for making the custom head fixation base and headplate.
‘Headfixation fixation base CAD.pdf’ is a mechanical drawing of the headfixation base, while ‘Head fixation base CAD.SLDPRT’ is the 3D drawing that can be opened in Solidworks software. ‘Head Plate CAD.pdf’ is a mechanical drawing of headplate implant, and ‘Head Plate CAD.SLDPRT’ is the 3D drawing that can be opened in Solidworks software. (ZIP 356 kb)
Supplementary Data 3: File archive consisting of mechanical drawings and computer aided design (CAD) files and instructions for assembling the autopatcher pipette actuator assembly.
Assembly instructions are provided in ‘Autopatcher Robotic Arm Assembly Manual.pdf’. ‘adapter plate 1.PDF’ and ‘adapter plate 1.SLDDRW’ are mechanical drawings of the adaptor plate used for mounting programmable linear stage onto Sutter manipulator. ‘adapter plate 1.SLDPRT’ is the corresponding 3D CAD file that can be opened in Solidworks. ‘adapter plate 2.PDF’ and ‘adapter plate 2.SLDDRW’ are mechanical drawings of the adaptor plate used to mount the amplifier headstage onto the programmable linear stage. ‘adapter plate 2.SLDPRT’ is the corresponding 3D CAD file that can be opened in Solidworks. (ZIP 2951 kb)
Supplementary Data 4: File archive consisting of mechanical drawings, computer aided design (CAD) files and instructions for assembling the autopatcher control box.
Assembly instructions are provided in the ‘Autopatcher control box assembly manual.pdf’ while ‘Autopatcher control box parts list.xlsx’ provides complete list of parts required for assembling the control box. Details of each part include description, name of vendor, catalog number, price/unit (as on Aug 2015), and quantity of each part. The sub-folder ‘Laser cutter files’ contains the ‘Autopatcher panels front & back.ai’ and ‘Autopatcher structural base, platform, & manometer clamp.ai’ files which can be used to cut two structural elements used for control box assembly (See the ‘Autopatcher control box assembly manual.pdf’). The sub-folder ‘Circuit board files’ contains: ‘Autopatcher PCB parts list.xlsx’ – a full parts list of all components on the pressure control printed circuit board (PCB) and valve relay PCB. ‘pressure_board.brd’ and ‘pressure_board.sch’ are the pressure control PCB CAD files, while ‘valve-relay_board.brd’ and ‘valve-relay_board.sch’ are the valve relay PCB CAD files. (ZIP 9892 kb)
Rights and permissions
About this article
Cite this article
Kodandaramaiah, S., Holst, G., Wickersham, I. et al. Assembly and operation of the autopatcher for automated intracellular neural recording in vivo. Nat Protoc 11, 634–654 (2016). https://doi.org/10.1038/nprot.2016.007
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.007
This article is cited by
-
A predictive model for seal condition in an automated patch clamp system
Journal of Micro and Bio Robotics (2022)
-
Automatic deep learning-driven label-free image-guided patch clamp system
Nature Communications (2021)
-
Craniobot: A computer numerical controlled robot for cranial microsurgeries
Scientific Reports (2019)
-
Automated identification of mouse visual areas with intrinsic signal imaging
Nature Protocols (2017)
-
In vivo whole-cell recording with high success rate in anaesthetized and awake mammalian brains
Molecular Brain (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.