Abstract
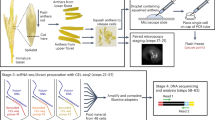
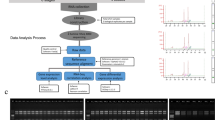
This protocol enables transcriptome profiling of specific cell or tissue types that are isolated from tomato using laser microdissection (LM). To prepare tissue for LM, fruit samples are first fixed in optimal cutting temperature (OCT) medium and frozen in molds. The tissue is then sectioned using a cryostat before being dissected using an LM instrument. The RNAs contained in the harvested cells are purified and subjected to two rounds of amplification to yield sufficient quantities of RNA to generate cDNA libraries. Unlike several other techniques that are used to isolate specific cell types, LM has the advantage of being readily applied to any plant species without having to generate transgenic plants. Using the protocols described here, LM-mediated cell-type transcriptomic analysis of two samples requires ∼8 d from tissue harvest to RNA sequencing (RNA-seq), whereas each additional sample, up to a total of 12 samples, requires ∼1 additional day for the LM step. RNA obtained using this method has been successfully used for deep-coverage transcriptome profiling, which is a particularly effective strategy for identifying genes that are differentially expressed between cell or tissue types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Martin, L.B.B., Fei, Z., Giovannoni, J.J. & Rose, J.K.C. Catalyzing plant science research with RNA-seq. Front. Plant Sci. doi: http://dx.doi.org/10.3389/fpls.2013.00066 (2013).
Hou, Z. et al. A cost effective RNA sequencing protocol for large-scale gene expression studies. Sci. Rep. 5, 9570 (2015).
Rogers, E.D., Jackson, T., Moussaieff, A., Aharoni, A. & Benfey, P.N. Cell type-specific transcriptional profiling: implications for metabolite profiling. Plant J. 70, 5–17 (2012).
Pattison, R.J. et al. Comprehensive tissue-specific transcriptome analysis reveals distinct regulatory programs during early tomato fruit development. Plant Physiol. 168, 1684–1701 (2015).
Qiao, Z. & Libault, M. Unleashing the potential of the root hair cell as a single plant cell type model in root systems biology. Front. Plant Sci. 4, 484 (2013).
Espina, V. et al. Laser-capture microdissection. Nat. Protoc. 1, 586–603 (2006).
Birnbaum, K. et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619 (2005).
Deal, R.B. & Henikoff, S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 6, 56–68 (2011).
Heiman, M., Kulicke, R., Fenster, R.J., Greengard, P. & Heintz, N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9, 1282–1291 (2014).
Brady, S.M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007).
Bargmann, B.O.R. & Birnbaum, K.D. Fluorescence activated cell sorting of plant protoplasts. J. Vis. Exp. 18, 1673 (2010).
Petersson, S.V., Lindén, P., Moritz, T. & Ljung, K. Cell-type specific metabolic profiling of Arabidopsis thaliana protoplasts as a tool for plant systems biology. Metabolomics 11, 1679–1689 (2015).
Gronlund, J.T., Eyres, A., Kumar, S., Buchanan-Wollaston, V. & Gifford, M.L. Cell specific analysis of Arabidopsis leaves using fluorescence activated cell sorting. J. Vis. Exp. 68, 4214 (2012).
Cheng, J. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154 (2005).
Barthelson, R.A., Lambert, G.M., Vanier, C., Lynch, R.M. & Galbraith, D.W. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics 8, 340 (2007).
Dinneny, J.R. et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945 (2008).
Chandran, D., Inada, N., Hather, G., Kleindt, C.K. & Wildermuth, M.C. Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site-specific processes and regulators. Proc. Natl. Acad. Sci. USA 107, 460–465 (2010).
Hacquart, S. et al. Laser capture microdissection of uredinia formed by Melampsora larici-populina revealed a transcriptional switch between biotrophy and sporulation. Mol. Plant Microbe Interact. 23, 1275–1286 (2010).
Gaude, N., Bortfeld, S., Duensing, N., Lohse, M. & Krajinski, F. Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J. 69, 510–528 (2012).
Hogekamp, C. & Küster, H. A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 10.1186/1471-2164-14-306 doi: http://dx.doi.org/10.1186/1471-2164-14-306 (2013).
Honaas, L.A. et al. Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 13 (2013).
Nakazono, M., Qiu, F., Borsuk, L.A. & Schnable, P.S. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15, 583–596 (2003).
Yu, P., Eggert, K., von Wirén, N., Li, C. & Hochholdinger, F. Cell type-specific gene expression analyses by RNA sequencing reveal local high nitrate-triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle regulation. Plant Physiol. 169, 690–704 (2015).
Matas, A.J., Agusti, J., Tadeo, F.R., Talon, M. & Rose, J.K.C. Tissue-specific transcriptome profiling of the citrus fruit epidermis and subepidermis using laser capture microdissection. J. Exp. Bot. 61, 3321–3330 (2010).
Matas, A.J. et al. Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23, 3893–3910 (2011).
Gao, C. et al. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol. J. 13, 370–382 (2015).
Wang, Y. et al. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol. 15, 548 (2014).
Xu, J. et al. An extensive proteome map of tomato (Solanum lycopersicum) fruit pericarp. Proteomics 13, 3059–3063 (2013).
Beisken, S. et al. Metabolic differences in ripening of Solanum lycopersicum 'Ailsa Craig' and three monogenic mutants. Sci. Data 1, 140029 (2014).
Perez-Fons, L. et al. A genome-wide metabolomic resource for tomato fruit from. Sci. Rep. 4, 3859 (2014).
Kerk, N.M., Ceserani, T., Tausta, S.L., Sussex, I.M. & Nelson, T.M. Laser capture microdissection of cells from plant tissues. Plant Physiol. 132, 27–35 (2003).
Goldsworthy, S.M., Stockton, P.S., Trempus, C.S., Foley, J.F. & Maronpot, R.R. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol. Carcinog. 25, 86–91 (1999).
Inada, N. & Wildermuth, M.C. Novel tissue preparation method and cell-specific marker for laser microdissection of Arabidopsis mature leaf. Planta 221, 9–16 (2005).
Takahashi, H. et al. A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. J. Plant Res. 123, 807–813 (2010).
Cai, S. & Lashbrook, C.C. Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling. Plant J. 48, 628–637 (2006).
Nelson, T., Tausta, S.L., Gandotra, N. & Liu, T. Laser microdissection of plant tissue: what you see is what you get. Annu. Rev. Plant Biol. 57, 181–201 (2006).
Golubeva, Y.G., Smith, R.M. & Sternberg, L.R. Optimizing frozen sample preparation for laser microdissection: assessment of CryoJane tape-transfer system®. PLoS One 8, e66854 (2013).
Vandewoestyne, M. et al. Laser capture microdissection: should an ultraviolet or infrared laser be used? Anal. Biochem. 439, 88–98 (2013).
Brooks, L. III et al. Microdissection of shoot meristem functional domains. PLoS Genet. 5, e1000476 (2009).
Clément-Ziza, M., Munnich, A., Lyonnet, S., Jaubert, F. & Besmond, C. Stabilization of RNA during laser capture microdissection by performing experiments under argon atmosphere or using ethanol as a solvent in staining solutions. RNA 14, 2698–2704 (2008).
Suarez-Quian, A.A., Tirado, O.M., Munell, F. & Reventos, J. Laser capture microdissection to assess development. In Methods of Enzymology vol. 356, Laser Capture Microscopy and Microdissection (ed. Conn, P.M.) (Elsevier Science, 2002).
Imbeaud, S. et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 33, 1–12 (2005).
Schoor, O. et al. Moderate degradation does not preclude microarray analysis of small amounts of RNA. BioTechniques 35, 1192–1201 (2003).
Gallego Romero, I., Pai, A.A., Tung, J. & Gilad, Y. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 12, 42 (2014).
Ladanyi, A et al. Laser microdissection in translational and clinical research. Cytometry A 69A, 947–960 (2006).
Nakamura, T. et al. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene 23, 2385–2400 (2004).
Turashvili, G. et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer 7, 55 (2007).
Kerman, I.A., Buck, B.J., Evans, S.J., Akil, H. & Watson, S.J. Combining laser capture microdissection with quantitative real-time PCR: effects of tissue manipulation on RNA quality and gene expression. J. Neurosci. Methods 153, 71–85 (2006).
Erickson, H.S. et al. Quantitative RT-PCR gene expression analysis of laser microdissected tissue samples. Nat. Protoc. 4, 902–922 (2009).
Kurimoto, K. & Saitou, M. Single-cell cDNA microarray profiling of complex biological processes of differentiation. Curr. Opin. Genet. Dev. 20, 470–477 (2010).
Emrich, S.J., Barbazuk, W.B., Li, L. & Schnable, P.S. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 17, 69–73 (2007).
Canas, R.A., Canales, J., Gomez-Maldonado, J., Avila, C. & Canovas, F.M. Transcriptome analysis in maritime pine using laser capture microdissection and 454 pyrosequencing. Tree Physiol. 34, 1278–1288 (2014).
Schad, M., Lipton, M.S., Giavalisco, P., Smith, R.D. & Kehr, J. Evaluation of two-dimentional electrophoresis and liquid chromatography-tandem mass spectrometry for tissue-specific protein profiling of laser-microdissected plant samples. Electrophoresis 26, 2729–2738 (2005).
Schad, M., Mungur, R., Fiehn, O. & Kehr, J. Metabolic profiling of laser microdissected vascular bundles of Arabidopsis thaliana. Plant Methods 1, 2 (2005).
Dembinsky, D. et al. Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 145, 575–588 (2007).
Ruel, K. et al. Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 189, 99–113 (2009).
Abbott, E., Hall, D., Hamberger, B. & Bohlmann, J. Laser microdissection of conifer stem tissues: isolation and analysis of high quality RNA, terpene synthase enzyme activity and terpenoid metabolites from resin ducts and cambial zone tissue of white spruce (Picea glauca). BMC Plant Biol. 10, 106 (2010).
Falter, C., Ellinger, D., von Hulsen, B., Heim, R. & Voigt, C.A. Simple preparation of plant epidermal tissue for laser microdissection and downstream quantitative proteome and carbohydrate analysis. Front. Plant Sci. doi: 10.3389/fpls.2015.00194 (2015).
Farlik, M. et al. Single-cell DNA methylome sequencing and bioinformatics inference of epigenomic cell-state dynamics. Cell Rep. 10, 1386–1397 (2015).
Acknowledgements
We acknowledge the Imaging Facility of Cornell University's Biotechnology Resource Center (Institute of Biotechnology) for the use of its Zeiss laser microdissection system. J.K.C.R. and C.C. were supported by a grant from the US National Science Foundation (Plant Genome Research Program; IOS-1339287), and Y.S. was partially supported by a Grant-in-Aid for JSPS Fellows from the Japan Society for the Promotion of Science (16J00582). A.J.M. was supported by the Spanish Ministry of Economy and Competitiveness with the Ramón y Cajal Fellowship (RYC-2011-08839).
Author information
Authors and Affiliations
Contributions
L.B.B.M., A.J.M., P.N. and Y.S. developed the protocol. L.B.B.M., P.N., C.C. and J.K.C.R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Annotation of cryostat features
a, block-holder; b, block in embedding mold; c, UV box; d, Object-holder; e, tightening screw; f, object-orientation knob; g, knife holder; h, angle control; i, anti-roll glass plate; j, CFSA 1X slide inside its protective wrapping; k, adhesive tape window; l, frame slide with PET-membrane; m, roller.
Supplementary information
Supplementary Text and Figures
Cryostat nomenclature. (PDF 266 kb)
Rights and permissions
About this article
Cite this article
Martin, L., Nicolas, P., Matas, A. et al. Laser microdissection of tomato fruit cell and tissue types for transcriptome profiling. Nat Protoc 11, 2376–2388 (2016). https://doi.org/10.1038/nprot.2016.146
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.146
This article is cited by
-
Laser Microdissection of Woody and Suberized Plant Tissues for RNA-Seq Analysis
Molecular Biotechnology (2023)
-
FIS1 encodes a GA2-oxidase that regulates fruit firmness in tomato
Nature Communications (2020)
-
Dissection of complex traits of tomato in the post-genome era
Theoretical and Applied Genetics (2020)
-
A protocol for laser microdissection (LMD) followed by transcriptome analysis of plant reproductive tissue in phylogenetically distant angiosperms
Plant Methods (2019)
-
Spatially resolved transcriptomics reveals plant host responses to pathogens
Plant Methods (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.