Abstract
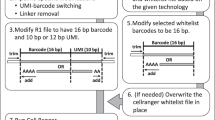
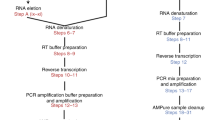
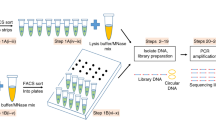
Parallel sequencing of a single cell's genome and transcriptome provides a powerful tool for dissecting genetic variation and its relationship with gene expression. Here we present a detailed protocol for G&T-seq, a method for separation and parallel sequencing of genomic DNA and full-length polyA(+) mRNA from single cells. We provide step-by-step instructions for the isolation and lysis of single cells; the physical separation of polyA(+) mRNA from genomic DNA using a modified oligo-dT bead capture and the respective whole-transcriptome and whole-genome amplifications; and library preparation and sequence analyses of these amplification products. The method allows the detection of thousands of transcripts in parallel with the genetic variants captured by the DNA-seq data from the same single cell. G&T-seq differs from other currently available methods for parallel DNA and RNA sequencing from single cells, as it involves physical separation of the DNA and RNA and does not require bespoke microfluidics platforms. The process can be implemented manually or through automation. When performed manually, paired genome and transcriptome sequencing libraries from eight single cells can be produced in ∼3 d by researchers experienced in molecular laboratory work. For users with experience in the programming and operation of liquid-handling robots, paired DNA and RNA libraries from 96 single cells can be produced in the same time frame. Sequence analysis and integration of single-cell G&T-seq DNA and RNA data requires a high level of bioinformatics expertise and familiarity with a wide range of informatics tools.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Shapiro, E., Biezuner, T. & Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 14, 618–630 (2013).
Kolodziejczyk, A.A., Kim, J.K., Svensson, V., Marioni, J.C. & Teichmann, S.A. The technology and biology of single-cell RNA sequencing. Mol. Cell 58, 610–620 (2015).
Wang, Y. & Navin, N.E. Advances and applications of single-cell sequencing technologies. Mol. Cell 58, 598–609 (2015).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014).
Voet, T. et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 41, 6119–6138 (2013).
Evrony, G.D. et al. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–496 (2012).
Evrony, G.D. et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron 85, 49–59 (2015).
Hou, Y. et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148, 873–885 (2012).
Xu, X. et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 148, 886–895 (2012).
Leung, M.L., Wang, Y., Waters, J. & Navin, N.E. SNES: single nucleus exome sequencing. Genome Biol. http://dx.doi.org/10.1186/s13059-015-0616-2 (2015).
de Bourcy, C.F. et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS One 9, e105585 (2014).
Hou, Y. et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. Gigascience 4, 37 (2015).
Huang, L., Ma, F., Chapman, A., Lu, S. & Xie, X.S. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu. Rev. Genomics Hum. Genet. 16, 79–102 (2015).
Gundry, M., Li, W., Maqbool, S.B. & Vijg, J. Direct, genome-wide assessment of DNA mutations in single cells. Nucleic Acids Res. 40, 2032–2040 (2012).
Wang, J., Fan, H.C., Behr, B. & Quake, S.R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412 (2012).
Macaulay, I.C. et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015).
Zong, C., Lu, S., Chapman, A.R. & Xie, X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 (2012).
Macaulay, I.C. & Voet, T. Single cell genomics: advances and future perspectives. PLoS Genet. 10, e1004126 (2014).
Hashimshony, T., Wagner, F., Sher, N. & Yanai, I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012).
Islam, S. et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat. Protoc. 7, 813–828 (2012).
Jaitin, D.A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Sasagawa, Y. et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA-Seq reveals non-genetic gene expression heterogeneity. Genome Biol. 14, R31 (2013).
Pan, X. et al. Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc. Natl. Acad. Sci. USA 110, 594–599 (2013).
Vanneste, E. et al. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 15, 577–583 (2009).
Dey, S.S., Kester, L., Spanjaard, B., Bienko, M. & van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 33, 285–289 (2015).
Li, W., Calder, R.B., Mar, J.C. & Vijg, J. Single-cell transcriptogenomics reveals transcriptional exclusion of ENU-mutated alleles. Mutat. Res. 772, 55–62 (2015).
Han, L. et al. Co-detection and sequencing of genes and transcripts from the same single cells facilitated by a microfluidics platform. Sci. Rep. 4, 6485 (2014).
Smallwood, S.A. et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014).
Angermueller, C. et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016).
Jiang, L. et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21, 1543–1551 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Quinlan, A.R. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.11–11.12.34 (2014).
Marcel, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 3 (2011).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Garvin, T. et al. Interactive analysis and assessment of single-cell copy-number variations. Nat. Methods 12, 1058–1060 (2015).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Anders, S., Pyl, P.T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Baslan, T. et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 25, 714–724 (2015).
Baslan, T. et al. Genome-wide copy number analysis of single cells. Nat. Protoc. 7, 1024–1041 (2012).
Nilsen, G. et al. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics 13, 591 (2012).
Cai, X. et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 8, 1280–1289 (2014).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
International HapMap, C. The International HapMap Project. Nature 426, 789–796 (2003).
Genomes Project, C. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Acknowledgements
This work was supported by the Wellcome Trust (grants to T.V. & C.P.P.) and funding from the Research Foundation Flanders (FWO) and the KU Leuven to T.V. (FWO–G.0687.12; KU Leuven SymBioSys, PFV/10/016). W.H. and C.P.P. were also funded by the Medical Research Council. M.J.T. is supported by a Wellcome Trust Sanger Institute Clinical PhD fellowship (UK).
Author information
Authors and Affiliations
Contributions
I.C.M., C.P.P. and T.V. devised and developed the method and wrote the manuscript. M.J.T. assisted with method development. W.H. and P.K. performed bioinformatics analysis of data.
Corresponding authors
Ethics declarations
Competing interests
T.V. is co-inventor on patent applications: WO/2011/157846, ‘Methods for haplotyping single cells’; WO/2014/053664, ‘High-throughput genotyping by sequencing’; and WO/2015/028576, ‘Haplotyping and copy number typing using polymorphic variant allelic frequencies’.
Integrated supplementary information
Supplementary Figure 1 Example robot deck layout for separation of DNA and RNA (steps 17-28)
Panel A shows the layout of the deck on the Beckman FxP robot for this process. Panel B shows a pictorial view of the deck layout with plates and tip types indicated.
Supplementary Figure 2 Example robot deck layout for separation of DNA and RNA (steps 47-55)
Panel A shows the layout of the deck on the Beckman FxP robot for this process. Panel B shows a pictorial view of the deck layout with plates and tip types indicated.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 314 kb)
Rights and permissions
About this article
Cite this article
Macaulay, I., Teng, M., Haerty, W. et al. Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nat Protoc 11, 2081–2103 (2016). https://doi.org/10.1038/nprot.2016.138
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.138
This article is cited by
-
Integrated transcriptomics and proteomics analysis reveals muscle metabolism effects of dietary Ulva lactuca and ulvan lyase supplementation in weaned piglets
Scientific Reports (2024)
-
Clonal hematopoiesis related TET2 loss-of-function impedes IL1β-mediated epigenetic reprogramming in hematopoietic stem and progenitor cells
Nature Communications (2023)
-
Benchmarking full-length transcript single cell mRNA sequencing protocols
BMC Genomics (2022)
-
Genomic insights into the phylogeny and biomass-degrading enzymes of rumen ciliates
The ISME Journal (2022)
-
Integrated Omics analysis of pig muscle metabolism under the effects of dietary Chlorella vulgaris and exogenous enzymes
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.