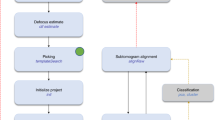
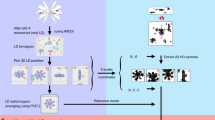
Abstract
Electron cryo-tomography (cryo-ET) is a technique that is used to produce 3D pictures (tomograms) of complex objects such as asymmetric viruses, cellular organelles or whole cells from a series of tilted electron cryo-microscopy (cryo-EM) images. Averaging of macromolecular complexes found within tomograms is known as subtomogram averaging, and this technique allows structure determination of macromolecular complexes in situ. Subtomogram averaging is also gaining in popularity for the calculation of initial models for single-particle analysis. We describe herein a protocol for subtomogram averaging from cryo-ET data using the RELION software (http://www2.mrc-lmb.cam.ac.uk/relion). RELION was originally developed for cryo-EM single-particle analysis, and the subtomogram averaging approach presented in this protocol has been implemented in the existing workflow for single-particle analysis so that users may conveniently tap into existing capabilities of the RELION software. We describe how to calculate 3D models for the contrast transfer function (CTF) that describe the transfer of information in the imaging process, and we illustrate the results of classification and subtomogram averaging refinement for cryo-ET data of purified hepatitis B capsid particles and Saccharomyces cerevisiae 80S ribosomes. Using the steps described in this protocol, along with the troubleshooting and optimization guidelines, high-resolution maps can be obtained in which secondary structure elements are resolved subtomogram.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Bai, X.C., McMullan, G. & Scheres, S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2015).
Cheng, Y. Single-particle cryo-EM at crystallographic resolution. Cell 161, 450–457 (2015).
Baumeister, W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr. Opin. Struct. Biol. 12, 679–684 (2002).
Briggs, J.A. Structural biology in situ—the potential of subtomogram averaging. Curr. Opin. Struct. Biol. 23, 261–267 (2013).
Beck, M., Lucić, V., Förster, F., Baumeister, W. & Medalia, O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 449, 611–615 (2007).
Grünewald, K. et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302, 1396–1398 (2003).
Briggs, J.A. et al. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. USA 106, 11090–11095 (2009).
Förster, F., Medalia, O., Zauberman, N., Baumeister, W. & Fass, D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 102, 4729–4734 (2005).
Nicastro, D. et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 (2006).
Schur, F.K. et al. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature 517, 505–508 (2015).
Pfeffer, S. et al. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 6, 8403 (2015).
Rigort, A. & Plitzko, J.M. Cryo-focused-ion-beam applications in structural biology. Arch. Biochem. Biophys. 581, 122–130 (2015).
Hrabe, T. et al. PyTom: a Python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J. Struct. Biol. 178, 177–188 (2012).
Bharat, T.A., Russo, C.J., Löwe, J., Passmore, L.A. & Scheres, S.H. Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure 23, 1743–1753 (2015).
Scheres, S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Scheres, S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Bharat, T.A. et al. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc. Natl. Acad. Sci. USA 111, 8233–8238 (2014).
Bharat, T.A. et al. Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy. Nature 487, 385–389 (2012).
Bartesaghi, A., Lecumberry, F., Sapiro, G. & Subramaniam, S. Protein secondary structure determination by constrained single-particle cryo-electron tomography. Structure 20, 2003–2013 (2012).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Heymann, J. & Belnap, D. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Castano-Diez, D., Kudryashev, M., Arheit, M. & Stahlberg, H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 178, 139–151 (2012).
Huiskonen, J.T. et al. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 84, 4889–4897 (2010).
Kremer, J.R., Mastronarde, D.N. & McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Crowther, R.A., Henderson, R. & Smith, J.M. MRC image processing programs. J. Struct. Biol. 116, 9–16 (1996).
Agulleiro, J.I. & Fernandez, J.J. Fast tomographic reconstruction on multicore computers. Bioinformatics 27, 582–583 (2011).
Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Hall, S.R. The star file–a new format for electronic data transfer and archiving. J. Chem. Inf. Comp. Sci. 31, 326–333 (1991).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Eibauer, M. et al. Unraveling the structure of membrane proteins in situ by transfer function corrected cryo-electron tomography. J. Struct. Biol. 180, 488–496 (2012).
Scheres, S.H. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife 3, e03665 (2014).
Yu, L., Snapp, R.R., Ruiz, T. & Radermacher, M. Projection-based volume alignment. J. Struct. Biol. 182, 93–105 (2013).
Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012).
Scheres, S.H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Rosenthal, P.B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Kucukelbir, A., Sigworth, F.J. & Tagare, H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Patwardhan, A. et al. A 3D cellular context for the macromolecular world. Nat. Struct. Mol. Biol. 21, 841–845 (2014).
Acknowledgements
We thank X.C. Bai, I.S. Fernandez and K. Vinothhumar for help with sample preparation; J. Grimmett and T. Darling for assistance with high-performance computing; S. Chen and C. Savva for assistance with electron microscopy; and J. Löwe for helpful discussions. This work was supported by funds from EMBO (ALTF 3-2013 and aALTF 778-2015 to T.A.M.B.) and the UK Medical Research Council (MC_UP_A025_1013 to S.H.W.S.).
Author information
Authors and Affiliations
Contributions
T.A.M.B. performed tomographic data acquisition and data processing, and developed the Python script for pre-processing of the subtomograms. S.H.W.S. developed the subtomogram procedures inside RELION. Both authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
The data set of seven tomograms of 80S ribosomes from S. cerevisiae has been deposited at the EMPIAR database (EMPIAR-10045), and the final map from 3D auto-refinement of these data has been submitted to the EMDB (EMD-3228).
Rights and permissions
About this article
Cite this article
Bharat, T., Scheres, S. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc 11, 2054–2065 (2016). https://doi.org/10.1038/nprot.2016.124
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.124
This article is cited by
-
Yersinia entomophaga Tc toxin is released by T10SS-dependent lysis of specialized cell subpopulations
Nature Microbiology (2024)
-
Plasma FIB milling for the determination of structures in situ
Nature Communications (2023)
-
Commercial influenza vaccines vary in HA-complex structure and in induction of cross-reactive HA antibodies
Nature Communications (2023)
-
A method for restoring signals and revealing individual macromolecule states in cryo-ET, REST
Nature Communications (2023)
-
Convolutional networks for supervised mining of molecular patterns within cellular context
Nature Methods (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.