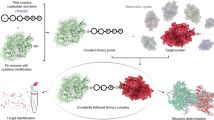
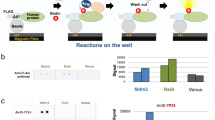
Abstract
AMPylation (adenylylation) has been recognized as an important post-translational modification that is used by pathogens to regulate host cellular proteins and their associated signaling pathways. AMPylation has potential functions in various cellular processes, and it is widely conserved across both prokaryotes and eukaryotes. However, despite the identification of many AMPylators, relatively few candidate substrates of AMPylation are known. This is changing with the recent development of a robust and reliable method for identifying new substrates using protein microarrays, which can markedly expand the list of potential substrates. Here we describe procedures for detecting AMPylated and auto-AMPylated proteins in a sensitive, high-throughput and nonradioactive manner. The approach uses high-density protein microarrays fabricated using nucleic acid programmable protein array (NAPPA) technology, which enables the highly successful display of fresh recombinant human proteins in situ. The modification of target proteins is determined via copper-catalyzed azide-alkyne cycloaddition (CuAAC). The assay can be accomplished within 11 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Stadtman, E.R. et al. Multiple molecular forms of glutamine synthetase produced by enzyme catalyzed adenylation and deadenylylation reactions. Adv. Enzyme Regul. 8, 99–118 (1970).
Itzen, A., Blankenfeldt, W. & Goody, R.S. Adenylylation: renaissance of a forgotten post-translational modification. Trends Biochem. Sci. 36, 221–228 (2011).
Yarbrough, M.L. et al. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272 (2009).
Worby, C.A. et al. The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34, 93–103 (2009).
Tan, Y. & Luo, Z.Q. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475, 506–509 (2011).
Neunuebel, M.R. et al. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333, 453–456 (2011).
Muller, M.P. et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949 (2010).
Kinch, L.N., Yarbrough, M.L., Orth, K. & Grishin, N.V. Fido, a novel AMPylation domain common to Fic, Doc, and AvrB. PLoS ONE 4, e5818 (2009).
Finn, R.D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Yu, X. et al. Copper-catalyzed azide-alkyne cycloaddition (click chemistry)-based detection of global pathogen-host AMPylation on self-assembled human protein microarrays. Mol. Cell. Proteomics 13, 3164–3176 (2014).
Woolery, A.R., Yu, X., LaBaer, J. & Orth, K. AMPylation of Rho GTPases subverts multiple host signaling processes. J. Biol. Chem. 289, 32977–32988 (2014).
Rahman, M. et al. Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat. Neurosci. 15, 871–875 (2012).
Ham, H. et al. Unfolded protein response-regulated dFic reversibly AMPylates BiP during endoplasmic reticulum homeostasis. J. Biol. Chem. 289, 36059–36069 (2014).
Pieles, K., Glatter, T., Harms, A., Schmidt, A. & Dehio, C. An experimental strategy for the identification of AMPylation targets from complex protein samples. Proteomics 14, 1048–1052 (2014).
Lewallen, D.M. et al. Inhibiting AMPylation: a novel screen to identify the first small molecule inhibitors of protein AMPylation. ACS Chem. Biol. 9, 433–442 (2014).
Lewallen, D.M., Steckler, C.J., Knuckley, B., Chalmers, M.J. & Thompson, P.R. Probing adenylation: using a fluorescently labelled ATP probe to directly label and immunoprecipitate VopS substrates. Mol. Biosyst. 8, 1701–1706 (2012).
Li, Y., Al-Eryani, R., Yarbrough, M.L., Orth, K. & Ball, H.L. Characterization of AMPylation on threonine, serine, and tyrosine using mass spectrometry. J. Am. Soc. Mass Spectrom. 22, 752–761 (2011).
Hao, Y.H. et al. Characterization of a rabbit polyclonal antibody against threonine-AMPylation. J. Biotechnol. 151, 251–254 (2011).
Grammel, M., Luong, P., Orth, K. & Hang, H.C. A chemical reporter for protein AMPylation. J. Am. Chem. Soc. 133, 17103–17105 (2011).
Ramachandran, N. et al. Self-assembling protein microarrays. Science 305, 86–90 (2004).
Ramachandran, N. et al. Next-generation high-density self-assembling functional protein arrays. Nat. Methods 5, 535–538 (2008).
Miersch, S. et al. Serological autoantibody profiling of type 1 diabetes by protein arrays. J. Proteomics 94, 486–496 (2013).
Prados-Rosales, R. et al. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 5, e01921 (2014).
Grammel, M. & Hang, H.C. Chemical reporters for biological discovery. Nat. Chem. Biol. 9, 475–484 (2013).
Westcott, N.P. & Hang, H.C. Chemical reporters for exploring ADP-ribosylation and AMPylation at the host-pathogen interface. Curr. Opin. Chem. Biol. 23C, 56–62 (2014).
Engel, P. et al. Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482, 107–110 (2012).
Bunney, T.D. et al. Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions. Structure 22, 1831–1843 (2014).
Yu, X. et al. Exploration of panviral proteome: high-throughput cloning and functional implications in virus-host interactions. Theranostics 4, 808–822 (2014).
Lin, Y.Y. et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136, 1073–1084 (2009).
Ptacek, J. et al. Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 (2005).
Wu, X. et al. Activation of diverse signalling pathways by oncogenic PIK3CA mutations. Nat. Commun. 5, 4961 (2014).
Yu, X., Schneiderhan-Marra, N. & Joos, T.O. Protein microarrays for personalized medicine. Clin. Chem. 56, 376–387 (2010).
Yu, X. et al. Quantifying antibody binding on protein microarrays using microarray nonlinear calibration. Biotechniques 54, 257–264 (2013).
Festa, F. et al. Robust microarray production of freshly expressed proteins in a human milieu. Proteomics Clin. Appl. 7, 372–377 (2013).
Qiu, J. & LaBaer, J. Nucleic acid programmable protein array a just-in-time multiplexed protein expression and purification platform. Methods Enzymol. 500, 151–163 (2011).
Miersch, S. & LaBaer, J. Nucleic acid programmable protein arrays: versatile tools for array-based functional protein studies. Curr. Protoc. Protein Sci. 64, 27.2.1–27.2.26 (2011).
Sibani, S. & LaBaer, J. Immunoprofiling using NAPPA protein microarrays. Methods Mol. Biol. 723, 149–161 (2011).
Seiler, C.Y. et al. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–D1260 (2014).
Garcia-Pino, A., Zenkin, N. & Loris, R. The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem. Sci. 39, 121–129 (2014).
Festa, F., Mendoza, A., Vatten, K. & Labaer, J. Study of the kinase activity using NAPPA protein microarray expressed with human IVTT system. Cancer Res. 72, LB-414 (2012).
Neunuebel, M.R., Mohammadi, S., Jarnik, M. & Machner, M.P. Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J. Bacteriol. 194, 1389–1400 (2012).
Kovacic, S. et al. Construction and characterization of kilobasepair densely labeled peptide-DNA. Biomacromolecules 15, 4065–4072 (2014).
Kim, S.Y., Kim, I.G., Chung, S.I. & Steinert, P.M. The structure of the transglutaminase 1 enzyme. Deletion cloning reveals domains that regulate its specific activity and substrate specificity. J. Biol. Chem. 269, 27979–27986 (1994).
Saul, J. et al. Development of a full-length human protein production pipeline. Protein Sci. 23, 1123–1135 (2014).
Carlson, E.D., Gan, R., Hodgman, C.E. & Jewett, M.C. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 30, 1185–1194 (2012).
Joshi, P. et al. The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol. 9, 672 (2013).
Spurrier, B., Ramalingam, S. & Nishizuka, S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat. Protoc. 3, 1796–1808 (2008).
Anderson, K.S. et al. Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J. Proteome Res. 10, 85–96 (2011).
Cruz, J.W. et al. Doc toxin is a kinase that inactivates elongation factor Tu. J. Biol. Chem. 289, 7788–7798 (2014).
Feng, F. et al. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485, 114–118 (2012).
Acknowledgements
We thank the Early Detection Research Network (5U01CA117374). We thank K. Orth's laboratory (Department of Molecular Biology, University of Texas Southwestern Medical Center) and H. Hang's laboratory (The Laboratory of Chemical Biology and Microbial Pathogenesis, The Rockefeller University) for providing the purified AMPylator proteins and click reagents, respectively. We thank B. Petritis and K. Barker for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
X.Y. designed and performed the experiments, and wrote the manuscript; J.L. designed the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Yu, X., LaBaer, J. High-throughput identification of proteins with AMPylation using self-assembled human protein (NAPPA) microarrays. Nat Protoc 10, 756–767 (2015). https://doi.org/10.1038/nprot.2015.044
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.044
This article is cited by
-
Legionella effector AnkX interacts with host nuclear protein PLEKHN1
BMC Microbiology (2018)
-
Self-assembling functional programmable protein array for studying protein–protein interactions in malaria parasites
Malaria Journal (2018)
-
Quantitative proteomics in lung cancer
Journal of Biomedical Science (2017)
-
The ORFeome Collaboration: a genome-scale human ORF-clone resource
Nature Methods (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.