Abstract
Tribolium castaneum has become an important insect model organism for evolutionary developmental biology, genetics and biotechnology. However, few protocols for live fluorescence imaging of Tribolium have been reported, and little image data is available. Here we provide a protocol for recording the development of Tribolium embryos with light-sheet–based fluorescence microscopy. The protocol can be completed in 4–7 d and provides procedural details for: embryo collection, microscope configuration, embryo preparation and mounting, noninvasive live imaging for up to 120 h along multiple directions, retrieval of the live embryo once imaging is completed, and image data processing, for which exemplary data is provided. Stringent quality control criteria for developmental biology studies are also discussed. Light-sheet–based fluorescence microscopy complements existing toolkits used to study Tribolium development, can be adapted to other insect species, and requires no advanced imaging or sample preparation skills.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout












Similar content being viewed by others
References
Klingler, M. Tribolium. Curr. Biol. 14, R639–R640 (2004).
Schröder, R., Beermann, A., Wittkopp, N. & Lutz, R. From development to biodiversity—Tribolium castaneum, an insect model organism for short germband development. Dev. Genes Evol. 218, 119–126 (2008).
Roth, S. & Hartenstein, V. Development of Tribolium castaneum. Dev. Genes Evol. 218, 115–118 (2008).
Brown, S.J. et al. The red flour beetle, Tribolium castaneum (Coleoptera): a model for studies of development and pest biology. Cold Spring Harb. Protoc. 4 10.1101/pdb.emo126 (2009).
Roth, S. & Panfilio, K.A. Development. Making waves for segments. Science 336, 306–307 (2012).
Richards, S. et al. The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955 (2008).
Shippy, T.D., Coleman, C.M., Tomoyasu, Y. & Brown, S.J. Concurrent in situ hybridization and antibody staining in red flour beetle (Tribolium) embryos. Cold Spring Harb. Protoc. 4, 968–973 (2009).
Berghammer, A.J., Weber, M., Trauner, J. & Klingler, M. Red flour beetle (Tribolium) germline transformation and insertional mutagenesis. Cold Spring Harb. Protoc. 4 doi:10.1101/pdb.prot5259 (2009).
Posnien, N. et al. RNAi in the red flour beetle (Tribolium). Cold Spring Harb. Protoc. 4 doi:10.1101/pdb.prot5256 (2009).
Sarrazin, A.F., Peel, A.D. & Averof, M. A segmentation clock with two-segment periodicity in insects. Science 336, 338–341 (2012).
El-Sherif, E., Averof, M. & Brown, S.J. A segmentation clock operating in blastoderm and germband stages of Tribolium development. Development 139, 4341–4346 (2012).
Benton, M.A., Akam, M. & Pavlopoulos, A. Cell and tissue dynamics during Tribolium embryogenesis revealed by versatile fluorescence labeling approaches. Development 140, 3210–3220 (2013).
Panfilio, K.A., Oberhofer, G. & Roth, S. High plasticity in epithelial morphogenesis during insect dorsal closure. Biol. Open 2, 1108–1118 (2013).
Koelzer, S., Kölsch, Y. & Panfilio, K.A. Visualizing late insect embryogenesis: extraembryonic and mesodermal enhancer trap expression in the beetle Tribolium castaneum. PLoS ONE 9, e103967 (2014).
Nollmann, F.I. et al. A photorhabdus natural product inhibits insect juvenile hormone epoxide hydrolase. Chembiochem 16, 766 (2015).
Nakamoto, A. et al. Changing cell behaviours during beetle embryogenesis correlates with slowing of segmentation. Nat. Commun. 6, 6635 (2015).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E.H.K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H.K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008).
Keller, P.J. et al. Fast, high-contrast imaging of animal development with scanned light-sheet–based structured-illumination microscopy. Nat. Methods 7, 637–642 (2010).
Strobl, F. & Stelzer, E.H.K. Non-invasive long-term fluorescence live imaging of Tribolium castaneum embryos. Development 141, 2331–2338 (2014).
Stelzer, E.H.K. & Lindek, S. Fundamental reduction of the observation volume in far-field light microscopy by detection orthogonal to the illumination axis: confocal theta microscopy. Opt. Commun. 111, 536–547 (1994).
Cox, I. Scanning optical fluorescence microscopy. J. Microsc. 133, 149–154 (1984).
Stelzer, E.H.K. Light-sheet fluorescence microscopy for quantitative biology. Nat. Methods 12, 23–26 (2015).
Hell, S., Reiner, G., Cremer, C. & Stelzer, E.H.K. Aberrations in confocal fluorescence microscopy induced by mismatches in refractive index. J. Microsc. 169, 391–405 (1993).
Pitrone, P.G. et al. OpenSPIM: an open-access light-sheet microscopy platform. Nat. Methods 10, 598–599 (2013).
Gualda, E.J. et al. OpenSpinMicroscopy: an open-source integrated microscopy platform. Nat. Methods 10, 599–600 (2013).
Huisken, J. & Stainier, D.Y.R. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 32, 2608–2610 (2007).
Krzic, U., Gunther, S., Saunders, T.E., Streichan, S.J. & Hufnagel, L. Multiview light-sheet microscope for rapid in toto imaging. Nat. Methods 9, 730–733 (2012).
Tomer, R., Khairy, K., Amat, F. & Keller, P.J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Tang, B., Wang, Y., Zhu, J. & Zhao, W. Web resources for model organism studies. Genomics Proteomics Bioinformatics 13, 64 (2015).
St Pierre, S.E., Ponting, L., Stefancsik, R. & McQuilton, P. FlyBase 102: advanced approaches to interrogating FlyBase. Nucleic Acids Res. 42, D780–D788 (2014).
Dos Santos, G. et al. FlyBase: introduction of the Drosophila melanogaster release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43, D690–D697 (2015).
Baldock, R.A. et al. EMAP and EMAGE: a framework for understanding spatially organized data. Neuroinformatics 1, 309–325 (2003).
Davidson, D., Bard, J., Kaufman, M. & Baldock, R. The mouse atlas database: a community resource for mouse development. Trends Genet. 17, 49–51 (2001).
Husz, Z.L., Burton, N., Hill, B., Milyaev, N. & Baldock, R.A. Web tools for large-scale 3D biological images and atlases. BMC Bioinformatics 13, 122 (2012).
Dönitz, J. et al. TrOn: an anatomical ontology for the beetle Tribolium castaneum. PLoS ONE 8, e70695 (2013).
Dönitz, J. et al. iBeetle-Base: a database for RNAi phenotypes in the red flour beetle Tribolium castaneum. Nucleic Acids Res. 43, D720–D725 (2014).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Gilles, A.F. & Averof, M. Functional genetics for all: engineered nucleases, CRISPR and the gene editing revolution. EvoDevo 5, 43 (2014).
Gillies, A.F. Schinko, J.B. & Averof, M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development 10.1242/dev.125054 (9 July 2015).
Bassett, A.R., Tibbit, C., Ponting, C.P. & Liu, J.-L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228 (2013).
Bassett, A.R. & Liu, J.-L. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genomics 41, 7–19 (2014).
Reynaud, E.G., Krzic, U., Greger, K. & Stelzer, E.H.K. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J. 2, 266–275 (2008).
Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H.K. Digital scanned laser light-sheet fluorescence microscopy (DSLM) of zebrafish and Drosophila embryonic development. Cold Spring Harb. Protoc. 6, 1235–1243 (2011).
Reynaud, E.G., Peychl, J., Huisken, J. & Tomancak, P. Guide to light-sheet microscopy for adventurous biologists. Nat. Methods 12, 30–34 (2014).
Khairy, K. & Keller, P.J. Reconstructing embryonic development. Genesis 49, 488–513 (2011).
Amat, F. & Keller, P.J. Towards comprehensive cell lineage reconstructions in complex organisms using light-sheet microscopy. Dev. Growth Differ. 55, 563–578 (2013).
Amat, F. et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11, 951–958 (2014).
Engelbrecht, C.J. & Stelzer, E.H.K. Resolution enhancement in a light-sheet-based microscope (SPIM). Opt. Lett. 31, 1477–1479 (2006).
Wu, Y. et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 17708–17713 (2011).
Kumar, A. et al. Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. Nat. Protoc. 9, 2555–2573 (2014).
Lorenzen, M.D. et al. piggyBac-mediated germline transformation in the beetle Tribolium castaneum. Insect Mol. Biol. 12, 433–440 (2003).
Trauner, J. et al. Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol. 7, 73 (2009).
Schinko, J.B. et al. Functionality of the GAL4/UAS system in Tribolium requires the use of endogenous core promoters. BMC Dev. Biol. 10, 53 (2010).
Siebert, K.S., Lorenzen, M.D., Brown, S.J., Park, Y. & Beeman, R.W. Tubulin-superfamily genes in Tribolium castaneum and the use of a tubulin promoter to drive transgene expression. Insect Biochem. Mol. Biol. 38, 749–755 (2008).
Lorenzen, M.D., Brown, S.J., Denell, R.E. & Beeman, R.W. Transgene expression from the Tribolium castaneum polyubiquitin promoter. Insect Mol. Biol. 11, 399–407 (2002).
Schinko, J.B., Hillebrand, K. & Bucher, G. Heat shock-mediated misexpression of genes in the beetle Tribolium castaneum. Dev. Genes Evol. 222, 287–298 (2012).
Terasaki, M. & Jaffe, L.A. Labeling of cell membranes and compartments for live cell fluorescence microscopy. Methods Cell Biol. 74, 469–489 (2004).
Kremers, G.-J., Gilbert, S.G., Cranfill, P.J., Davidson, M.W. & Piston, D.W. Fluorescent proteins at a glance. J. Cell Sci. 124, 157–160 (2011).
Shaner, N.C., Steinbach, P.A. & Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 (2005).
Chudakov, D.M., Matz, M.V., Lukyanov, S. & Lukyanov, K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163 (2010).
Kaufmann, A., Mickoleit, M., Weber, M. & Huisken, J. Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development 139, 3242–3247 (2012).
Ichikawa, T. et al. Live imaging of whole mouse embryos during gastrulation: migration analyses of epiblast and mesodermal cells. PLoS ONE 8, e64506 (2013).
Ichikawa, T. et al. Live imaging and quantitative analysis of gastrulation in mouse embryos using light-sheet microscopy and 3D tracking tools. Nat. Protoc. 9, 575–585 (2014).
Breuninger, T., Greger, K. & Stelzer, E.H.K. Lateral modulation boosts image quality in single plane illumination fluorescence microscopy. Opt. Lett. 32, 1938–1940 (2007).
Swoger, J., Verveer, P., Greger, K., Huisken, J. & Stelzer, E.H.K. Multi-view image fusion improves resolution in three-dimensional microscopy. Opt. Express 15, 8029–8042 (2007).
Preibisch, S., Saalfeld, S., Schindelin, J. & Tomancak, P. Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010).
Preibisch, S. et al. Efficient Bayesian-based multiview deconvolution. Nat. Methods 11, 645–648 (2014).
Walter, T. et al. Visualization of image data from cells to organisms. Nat. Methods 7, S26–S41 (2010).
Stelzer, E.H.K. Contrast, resolution, pixelation, dynamic range and signal-to-noise ratio: fundamental limits to resolution in fluorescence light microscopy. J. Microsc. 189, 15–24 (1998).
Keller, P.J. & Stelzer, E.H.K. Digital scanned laser light sheet fluorescence microscopy. Cold Spring Harb. Protoc. 5 10.1101/pdb.top78 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Greger, K., Swoger, J. & Stelzer, E.H.K. Basic building units and properties of a fluorescence single plane illumination microscope. Rev. Sci. Instrum. 78, 023705 (2007).
Arpino, J.A.J., Rizkallah, P.J. & Jones, D.D. Crystal structure of enhanced green fluorescent protein to 1.35 Å resolution reveals alternative conformations for Glu222. PLoS ONE 7, e47132 (2012).
Sokoloff, A. The Biology of Tribolium: With Special Emphasis on Genetic Aspects (Oxford University Press, 1974).
Kittler, J. & Illingworth, J. Minimum error thresholding. Pattern Recogn. 19, 41–47 (1986).
Cho, S., Haralick, R. & Yi, S. Improvement of Kittler and Illingworth's minimum error thresholding. Pattern Recogn. 22, 609–617 (1989).
Acknowledgements
We thank A. Peel (University of Leeds) and M. Averof (Institut de Génomique Fonctionnelle de Lyon) for the Tribolium EFA-nGFP line; D. Kleinhans for kindly providing a raw version of the mDSLM principle scheme; D. von Wangenheim for help with the generation of the videos; S. Plath and K. Schmale for technical assistance; and J. Alexander Ross, A. Anderl and S. Fischer for their general support. The research was funded by the Cluster of Excellence Frankfurt for Macromolecular Complexes (CEF-MC, EXC 115) granted to the Buchmann Institute for Molecular Life Sciences (BMLS) at the Goethe University—Frankfurt am Main by the Deutsche Forschungsgemeinschaft (DFG).
Author information
Authors and Affiliations
Contributions
All authors conceived the research and wrote the publication. F.S. performed the experiments to obtain the example results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Principle effects of LSFM illumination and detection on image quality.
(A) Top view of a scheme showing the arrangement of illumination objective, sample chamber and detection objective in a light sheet-based fluorescence microscope. The inset depicts the illumination and detection of a single line in a specimen. (B) Top view of the objective arrangement in the mDSLM. In the inset, image quality indicators are provided as colored arcs. The green arc marks the quarter with an expected high quality, the orange arcs the quarters with an expected medium quality and the red arc the quarter with an expected low quality. (C) Y maximum projection of a Z stack with the image quality indicators introduced in B.
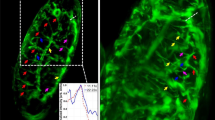
Supplementary Figure 2 Example quantification of the serosa window area over time during serosa window closure.
(A) Z maximum projection of a Tribolium embryo during gastrulation that commences serosa window closure seen from the ventral side. (B) Time lapse of single planes, 26 µm below the ventral surface of the embryo, showing the process of serosa window closure. The first row depicts the raw planes. In the second row, an entropy filter was applied to the raw planes. Then, an intensity threshold was set to obtain the binary images. The binary images were inverted and further processed by morphological operations to extract the serosa window area in each image, as shown in the third row. The fourth row illustrates the raw planes marked with the extracted serosa window area. (C) A plot of the serosa window area as a function of development time, providing evidence that the serosa window area during serosa window closure follows an exponential decay. ZP, Z maximum projection with image processing; PA, plane with intensity adjustment.
Supplementary Figure 3 Chronological staging table of Tribolium embryogenesis.
The left column contains Z maximum projections along four orientations (0° (ventral), 90° (lateral), 180° (dorsal), and 270° (lateral)) of a Tribolium embryo at the beginning of five major embryogenetic events. In the middle column, an outline of the most important developmental processes with a temporal indication in hours (developments at 35°C and at room temperature) and stage indication in percent of full development. Please note that 00:00 h relates to our standard imaging starting point (that is after a 1 h egg laying period / 15 h of incubation at 25°C and 1 h of preparation at room temperature), whereas the stage indication in percent is provided for the whole of embryogenesis. The right column shows Z maximum projections of developmental processes during the respective embryogenetic events that have been the focus of previous research. Red arrows indicate the location of the process mentioned in the bullet points. RT, room temperature (23±1°C); ap., approximately.
Supplementary Figure 4 Scheme of the recommended control experiments.
In addition to the imaging procedure of the fluorescent embryo (left column), we suggest two control experiments that can be run in parallel: a fluorescent control embryo (middle column) and a wild type control embryo (right column). All steps in which the control experiments have to be considered are indicated and also mentioned in the Procedure section.
Supplementary Figure 5 Operating principle of the digital scanned laser light sheet-based fluorescence microscope (DSLM).
In contrast to other types of light sheet-based fluorescence microscopes, the DSLM does not have a static light sheet. Instead, a dynamic light sheet is generated by scanning the laser beam with a piezo-driven two-axes scanning mirror (see green annotations). The laser beam, originating from the laser source, is guided by the scanning mirror through an f-theta and a tube lens into the illumination objective. The dynamic light sheet illuminates a thin slice of the Tribolium embryo, and the emitted fluorescence is guided by a perpendicularly arranged detection objective through an appropriate filter and a tube lens into the camera. The detail on the right shows the sample holder with the embryo in front of the detection objective. Adapted and modified from ref. 18 with permission, Nature Publishing Group.
Supplementary Figure 6 Perfusion system.
(A) Overview of mDSLM with connected perfusion system. The main elements of the circulatory perfusion system are labelled. Controlled transfer of PBS from the reservoir is achieved via the inflow pump through the heating tube into the sample chamber. From the chamber, excessive PBS is transferred via the outflow pump back into the reservoir. Upper detail box shows a top view of the sample chamber. The perfusion system connects to the periphery of the sample chamber and, therefore, does not impair the imaging process. Lower detail box shows the front view of the sample chamber with connected inflow and outflow tubes. The inflow tube connects to the lower and the outflow tube to the upper joint of the sample chamber. Therefore, the PBS outflow rate is limited by the inflow rate. Directions of flow are indicated with white arrowheads. A third tube is connected to a catch basin located beneath the sample chamber in case of a leakage, as indicated with a gray arrowhead. (B) Setup and equilibrium of perfusion system. A constant temperature in the sample chamber is maintained via a combination of PBS flow rate and the temperature generated in the heating tube. Prior to the experiment, the perfusion system is given two hours to ensure a stable temperature in the sample chamber. The circulatory flow of PBS and relative warmth are indicated with colored arrowheads.
Supplementary Figure 7 Illustration of equipment setup.
(A) Front (left) and side view (right) of the glass cover slip holder. (A’) Front (left panel) and side view (right panel) of the holder in A with attached glass cover slip. (A’’) Glass cover slip holder in the LSFM sample chamber. From left to right: top view of glass cover slip holder perpendicular to Z (the detection axis), top, front and side view of the glass cover slip holder rotated by 45° around Y (the rotation axis). (B) Capillary holder (left panel) with capillary attached (right panel). (C) Syringe components with cut lines indicated in orange (left panel). Components with indicated parts cut off (right panel). (C’) Assembled syringe (top panel), with rubber hose (middle panel) and capillary (bottom panel) attached. (D) Humidity chamber. (D’) Scaffold created from serological pipette fragments without (left panel) and with object slide (right panel).
Supplementary Figure 8 Test recording of agarose-embedded beads column for microscope calibration.
The agarose-embedded beads column is shown along four orientations as a Z (first row), X (second row) and Y (third row) maximum projections. The maximum projections illustrate correct calibration of the microscope without any imaging artifacts. The rotated Y maximum projections (fourth row) demonstrate a correctly set up recording of the bead agarose column. Individual beads can be identified along all directions as shown in the detail images (fifth row). Due to the point spread function, beads are expected to appear as bright, round dots in the Z maximum projection and elongated along X and Y maximum projections. ZP, Z maximum projections with image processing; XP, X maximum projections with image processing; YP, Y maximum projections with image processing.
Supplementary Figure 9 Test recording of the embryo along four directions prior to time lapse imaging.
The embryo is shown in four orientations in the transmission light channel (first row) and as Z (second row), X (third row) and Y (fourth row) maximum projections. The maximum projections illustrate a correctly set up recording along four directions without any imaging artifacts. Some preparation issues and misconfigurations of the imaging setup (e.g. insufficient amount of agarose in the stability layer leading to a drift of the embryo, incorrect Z spacing, incorrect configured volume of view), might be more apparent in X and Y maximum projections than in the Z maximum projections. Indicators for a correctly set up recording are: X and Y maximum projections depict smooth object boundaries and show no motion blur; Z maximum projections and the corresponding X maximum projections exhibit high similarity; and in the Y maximum projections, the shape of the embryo should be roughly circular. ZP, Z maximum projections with image processing; XP, X maximum projections with image processing; YP, Y maximum projections with image processing.
Supplementary Figure 10 Qualitative coverage of the embryo.
The embryo is shown along four orientations as Z and Y maximum projections. The Y maximum projections (second row) illustrate that the quality of the recording strongly depends on the amount and type of biological material passed by the illumination and emission light paths. If no biological material obstructs the light paths, the resulting image quality is high (lower left area in all orientations). However, if the amount of material that has to be passed increases, the image quality drops (upper left and lower right areas in all orientations). The image quality is worst in areas where both light paths have to pass the complete embryo (upper right areas in all orientations). The rotated Y maximum projections (third row) highlight the fact that features of the embryo might only be partly visible in some orientations. The green lines outline the clearly detectable germ band borders from the respective direction. ZP, Z maximum projections with image processing; YP, Y maximum projections with image processing.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 – 10, Supplementary Table 1 and Supplementary Note (PDF 3767 kb)
Supplementary Script 1
Custom script 'Strobl2015A - ProCroRo.ijm' for the software FIJI for rotation, cropping, and computation of Z maximum projections in batch mode. The script can be downloaded at http://www.physikalischebiologie.de/bugcube (ZIP 1 kb)
Supplementary Script 2
Custom script 'Strobl2015 – BackCorMeanTra.m' for the software Mathematica to perform background correction and mean intensity transformation as described in Box 2. The script can be downloaded at http://www.physikalischebiologie.de/bugcube. (ZIP 5 kb)
Tribolium 90° (lateral) optical sectioning at two time points from −26.0 μm to 309.4 μm with a spacing of 2.58 μm between single planes.
Please note that the video is not rotated and the anterior pole of the embryo is at the bottom. The embryo exhibits active movement at approximately 145.6 μm optical sectioning depth; PA, single plane with intensity adjustment. (AVI 758 kb)
Tribolium 90° (lateral) real-time transmission light imaging.
Please note that the video is not rotated and the anterior pole of the embryo is at the bottom. The embryo stretches forcefully and then returns to the original position again. After the real-time video, a 5× slow motion of the stretching process is shown. (AVI 663 kb)
Tribolium embryogenesis along four orientations from 00:00 h to 120:00 h with an interval of 00:30 h between the time points.
The video starts with the rearrangement of the blastoderm and ends once dorsal closure is completed, showing four fundamental embryogenetic events of the insect class: gastrulation, germ band elongation, germ band retraction and dorsal closure. Frame rate is five frames per second. MP, maximum projection with image processing. (AVI 5408 kb)
Tribolium optical sectioning along four orientations from −26.0 μm to 309.4 μm with a spacing of 2.58 μm between single planes.
The first single plane with moderate signal was defined as 0 μm. The red line in the Z maximum projection indicates where the currently shown 0° (ventral) single plane is located. Frame rate is ten frames per second. ZP, Z maximum projection with image processing; PA, single plane with intensity adjustment. (AVI 3983 kb)
Tribolium 90° (lateral) optical sectioning at three time points from −26.0 μm to 309.4 μm with a spacing of 2.58 μm between single planes.
The first single plane with moderate signal was defined as 0 μm. The red line in the 90° (lateral) maximum projection indicates where the currently shown 0° (ventral) single plane is located. Frame rate is ten frames per second. ZP, Z maximum projection with image processing; PA, single plane with intensity adjustment. (AVI 3016 kb)
Tribolium 90° (lateral) optical sectioning shown in uniform and structured illumination light sheet mode from −26.0 μm to 309.4 μm with a spacing of 2.58 μm between single planes.
The first single plane with a moderate signal was defined as 0 μm. The red line in the 90° Turn Z maximum projection indicates where the currently shown 90° (lateral) single plane is located. The structured illumination mode reduces the blur from out-of-focus regions. Frame rate is ten frames per second. PA, single plane with intensity adjustment. (AVI 2074 kb)
Rights and permissions
About this article
Cite this article
Strobl, F., Schmitz, A. & Stelzer, E. Live imaging of Tribolium castaneum embryonic development using light-sheet–based fluorescence microscopy. Nat Protoc 10, 1486–1507 (2015). https://doi.org/10.1038/nprot.2015.093
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.093
This article is cited by
-
In toto light sheet fluorescence microscopy live imaging datasets of Ceratitis capitata embryonic development
Scientific Data (2022)
-
A deterministic genotyping workflow reduces waste of transgenic individuals by two-thirds
Scientific Reports (2021)
-
Light sheet fluorescence microscopy
Nature Reviews Methods Primers (2021)
-
Ultra-thin fluorocarbon foils optimise multiscale imaging of three-dimensional native and optically cleared specimens
Scientific Reports (2019)
-
Multiscale imaging of plant development by light-sheet fluorescence microscopy
Nature Plants (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.