Abstract
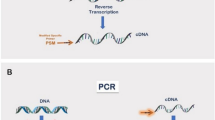
The 5′ ends of transcripts provide important information about transcription initiation sites and the approximate locations of local cis-acting enhancer elements; it is therefore important to establish the 5′ ends with some precision. RACE (rapid amplification of cDNA ends) PCR is useful for quickly obtaining full length cDNAs for mRNAs for which only part of the sequence is known and to identify alternative 5′ or 3′ ends of fully sequenced genes. The method consists of using PCR to amplify, from complex mixtures of cellular mRNA, the regions between the known parts of the sequence and non-specific tags appended to the ends of the cDNA. Whereas the poly(A) tail serves to provide such a tag at the 3′ end of the mRNA, an artificial one needs to be generated at the 5′ end, and various approaches have been described to address this step. The classical scheme for 5′ RACE described here is simple, suffices in many instances in which RACE is needed and can be performed in 1–3 days.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Frohman, M.A., Dush, M.K. & Martin, G.R. Rapid production of full-length cDNAs from rare transcripts by amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002 (1988).
Loh, E.L., Elliott, J.F., Cwirla, S., Lanier, L.L. & Davis, M.M. Polymerase chain reaction with single sided specificity: analysis of T cell receptor δ chain. Science 243, 217–220 (1989).
Ohara, O., Dorit, R.I. & Gilbert, W. One-sided PCR: the amplification of cDNA. Proc. Natl. Acad. Sci USA. 86, 5673–5677 (1989).
Frohman, M.A. & Martin, G.R. Rapid amplification of cDNA ends using nested primers. Tech. 1, 165–173 (1989).
Frohman, M.A. In PCR Protocols and Applications: A Laboratory Manual (eds. Innis, M., Gelfand, D., Sninsky, J. and White, T.) 28–38 (1989).
Frohman, M.A. Rapid amplification of cDNA for generation of full-length cDNA ends: thermal RACE. Methods Enzymol. 218, 340–356 (1993).
Frohman, M.A. In PCR. The Polymerase Chain Reaction. Methods in Molecular Biology Series (eds. Mullis, K. B., Ferre, F. & Gibbs, R. A.) 14–37 (Humana Press, Totowa, New Jersey, 1994).
Bertling, W.M., Beier, F. & Reichenberger, E. Determination of 5′ ends of specific mRNAs by DNA Ligase-dependent amplification. PCR Methods Applic. 3, 95–99 (1993).
Borson, N.D., Salo, W.L. & Drewes, L.R. A lock-docking oligo(dT) primer for 5′ and 3′ RACE PCR. PCR Methods Applic. 2, 144–148 (1992).
Dumas, J.B., Edwards, M., Delort, J. & Mallet, J. Oligodeoxyribonucleotide ligation to single-stranded cDNAs: a new tool for cloning 5′ ends of mRNAs and for constructing cDNA libraries by in vitro amplification. Nucleic Acids Res. 19, 5227–5233 (1991).
Fritz, J.D., Greaser, M.L. & Wolff, J.A. A novel 3′ extension technique using random primers in RNA-PCR. Nucleic Acids Res. 119, 3747 (1991).
Jain, R., Gomer, R.H. & Murtagh, J.J.J. Increasing specificity from the PCR-RACE technique. BioTechniques 12, 58–59 (1992).
Monstein, H.J., Thorup, J.U., Folkesson, R., Johnsen, A.H. & Rehfeld, J.F. cDNA deduced procionin — structure and expression in protochordates resemble that of procholecystokinin in mammals. FEBS Letters 331, 60–64 (1993).
Rashtchian, A., Buchman, G.W., Schuster, D.M. & Berninger, M.S. Uracil DNA glycosylase-mediated cloning of PCR-amplified DNA: application to genomic and cDNA cloning. Anal. Biochem. 206, 91–97 (1992).
Templeton, N.S., Urcelay, E. & Safer, B. Reducing artifact and increasing the yield of specific DNA target fragments during PCR-RACE or anchor PCR. BioTechniques 15, 48–50 (1993).
Scotto–Lavino, E., Du, G. & Frohman, M.A. 3′ end cDNA amplification using classic RACE. Nature Protocols (2006), doi: 10.1038/nprot.2006.481.
Fromont–Racine, M., Bertrand, E., Pictet, R. & Grange, T. A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res. 21, 1683–1684 (1993).
Bertrand, E., Fromont–Racine, M., Pictet, R. & Grange, T. Visualization of the interaction of a regulatory protein with RNA in vivo. Proc. Natl. Acad. Sci. USA 90, 3496–3500 (1993).
Brock, K.V., Deng, R. & Riblet, S.M. Nucleotide sequencing of 5′ and 3′ termini of bovine viral diarrhea virus by RNA ligation and PCR. Virol. Methods 38, 39–46 (1992).
Liu, X. & Gorovsky, M.A. Mapping the 5′ and 3′ ends of tetrahymena-thermophila mRNAs using RNA Ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res. 21, 4954–4960 (1993).
Mandl, C.W., Heinz, F.X., Puchhammer–Stockl, E. & Kunz, C. Sequencing the termini of capped viral RNA by 5′–3′ ligation and PCR. BioTechniques 10, 484–486 (1991).
Sallie, R. Characterization of the extreme 5′ ends of RNA molecules by RNA ligation-PCR. PCR Methods Applic. 3, 54–56 (1993).
Tessier, D.C., Brousseau, R. & Vernet, T. Ligation of single-stranded oligodeoxyribonucleotides by T4 RNA ligase. Anal. Biochem. 158, 171–178 (1986).
Volloch, V., Schweitzer, B., Zhang, X. & Rits, S. Identification of negative-strand complements to cytochrome oxidase subunit III RNA in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 88, 10671–10675 (1991).
Scotto–Lavino, E., Du, G. & Frohman, M.A. 5′ end cDNA amplification using new RACE. Nat. Protocols (2006), doi: 10.1038/nprot.2006.479.
Schmidt, W.M. & Mueller, M.W. CapSelect: a highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 27, e31 (1999).
Schramm, G., Bruchhaus, I. & Roeder, T. A simple and reliable 5′-RACE approach. Nucleic Acids Res. 28, e96 (2000).
Huang, J.C. & Chen, F. Simultaneous amplification of 5′ and 3′ cDNA ends based on template-switching effect and inverse PCR. Biotechniques 40, 187–189 (2006).
Mica, E., Gianfranceschi, L. & Pe, M.E. Characterization of five microRNA families in maize. J. Exp. Bot. 57, 2601–2612 (2006).
Wang, X.J., Reyes, J.L., Chua, N.H. & Gaasterland, T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5, r65 (2004).
Schnoor, M. et al. Characterization of the synthetic compatible solute homoectoine as a potent PCR enhancer. Biochem. Biophys. Res. Commun. 322, 867–872 (2004).
Shi, X. & Jarvis, D.L. A new rapid amplification of cDNA ends method for extremely guanine plus cytosine-rich genes. Anal. Biochem. 356, 222–228 (2006).
Sambrook, J., Fritsch, E.F. & Maniatis, T. In Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1989).
Sarker, G., Kapelner, S. & Sommer, S.S. Formamide can dramatically improve the specificity of PCR. Nucleic Acids Res. 18, 7465 (1990).
Zhang, Y. RACE all the way to the end. in Generation of cDNA libraries (ed. Ying, S.-Y.) 13–24 (Humana Press, Totow1a, NJ, 2003).
Acknowledgements
The author thanks Yue Zhang and the publisher for permission to use and adapt material from 'RACE all the way to the end' (ref. 35). This work was supported by awards NIHDDK 64166 and NIHGM71520 to M.A.F., and NIHGM071475 and a Scientist Development Grant from the American Heart Association to G.D. (0430096N).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Scotto–Lavino, E., Du, G. & Frohman, M. 5′ end cDNA amplification using classic RACE. Nat Protoc 1, 2555–2562 (2006). https://doi.org/10.1038/nprot.2006.480
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.480
This article is cited by
-
Downregulation of hepatic lncRNA Gm19619 improves gluconeogenesis and lipogenesis following vertical sleeve gastrectomy in mice
Communications Biology (2023)
-
MG1 interacts with a protease inhibitor and confers resistance to rice root-knot nematode
Nature Communications (2023)
-
T-cell and B-cell repertoire diversity are selectively skewed in children with idiopathic nephrotic syndrome revealed by high-throughput sequencing
World Journal of Pediatrics (2023)
-
Heterologous expression of Sesuvium portulacastrum SOS-related genes confer salt tolerance in yeast
Acta Physiologiae Plantarum (2023)
-
A versatile 5′ RACE-Seq methodology for the accurate identification of the 5′ termini of mRNAs
BMC Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.