Abstract
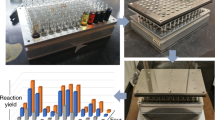
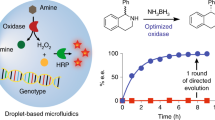
A procedure for the high-throughput screening of esterases is described. This includes enzyme expression in microtiter plates and the measurement of activity and enantioselectivity (E) of the esterase variants using acetates of secondary alcohols as model substrates. Acetic acid released is converted in an enzyme cascade leading to the stoichiometric formation of NADH, which is quantified in a spectrophotometer. The method allows screening of several thousand mutants per day and has already been successfully applied to identify an esterase mutant with an E>100 toward an important building block for organic synthesis. This protocol can also be used for lipases and possibly other hydrolases that are expressed in soluble form in conventional Escherichia coli strains. This protocol can be completed in 3–4 days.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bornscheuer, U.T. & Kazlauskas, R.J. Hydrolases in Organic Synthesis—Regio- and Stereoselective Biotransformations 2nd edn. (Wiley-VCH, Weinheim, 2005).
Lorenz, P., Liebeton, K., Niehaus, F. & Eck, J. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr. Opin. Biotechnol. 13, 572–577 (2002).
Handelsman, J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685 (2004).
Cherry, J.R. & Fidantsef, A.L. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 14, 438–443 (2003).
Turner, N.J. Directed evolution of enzymes for applied biocatalysis. Trends Biotechnol. 21, 474–478 (2003).
Schmidt, M., Baumann, M., Henke, E., Konarzycka-Bessler, M. & Bornscheuer, U.T. Directed evolution of lipases and esterases. Methods Enzymol. 388, 199–207 (2004).
Wahler, D. & Reymond, J.-L. High-throughput screening for biocatalysts. Curr. Opin. Chem. Biol. 12, 535–544 (2001).
Reetz, M.T. Controlling the enantioselectivity of enzymes by directed evolution: practical and theoretical ramnifications. Proc. Natl. Acad. Sci. USA 101, 5716–5722 (2004).
Goddard, J.P. & Reymond, J.-L. Recent advances in enzyme assays. Trends Biotechnol. 22, 363–370 (2004).
Reymond, J.-L. Enzyme assays. (Wiley-VCH, Weinheim, 2005).
Schmidt, M. & Bornscheuer, U.T. High-throughput assays for lipases and esterases. Biomol. Eng. 22, 51–56 (2005).
Reetz, M.T. & Jaeger, K.E. Enantioselective enzymes for organic synthesis created by directed evolution. Chem. Eur. J. 6, 407–412 (2000).
Schmidt, M. et al. Directed evolution of an esterase from Pseudomonas fluorescens yields a mutant with excellent enantioselectivity and activity for the kinetic resolution of a chiral building block. ChemBioChem 7, 805–809 (2006).
Henke, E. & Bornscheuer, U.T. Directed evolution of an esterase from Pseudomonas fluorescens. Random mutagenesis by error-prone PCR or a mutator strain and identification of mutants showing enhanced enantioselectivity by a resorufin-based fluorescence assay. Biol. Chem. 380, 1029–1033 (1999).
Baumann, M., Stürmer, R. & Bornscheuer, U.T. A high-throughput-screening method for the identification of active and enantioselective hydrolases. Angew. Chem. Int. Ed. 40, 4201–4204 (2001).
Leung, D.W., Chen, E. & Goeddel, D.V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1, 11–15 (1989).
Arnold, F.H. & Georgiou, G. (eds.) Methods in Molecular Biology, Vol.230: Directed Enzyme Evolution: Methods and Protocols Vol. 2 (Humana Press, Totawa, 2003).
Chung, C.T., Niemela, S.L. & Miller, R.H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86, 2172–2175 (1989).
Acknowledgements
We thank M. Baumann, E. Henke and M. Schmidt for their support in developing and applying the methodology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Böttcher, D., Bornscheuer, U. High-throughput screening of activity and enantioselectivity of esterases. Nat Protoc 1, 2340–2343 (2006). https://doi.org/10.1038/nprot.2006.391
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.391
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.