Abstract
Mass spectrometry imaging is a powerful tool for directly determining the distribution of proteins, peptides, lipids, neurotransmitters, metabolites and drugs in neural tissue sections in situ. Molecule-specific imaging can be achieved using various ionization techniques that are suited to different applications but which all yield data with high mass accuracies and spatial resolutions. The ability to simultaneously obtain images showing the distributions of chemical species ranging from metal ions to macromolecules makes it possible to explore the chemical organization of a sample and to correlate the results obtained with specific anatomical features. The imaging of biomolecules has provided new insights into multiple neurological diseases, including Parkinson’s and Alzheimer’s disease. Mass spectrometry imaging can also be used in conjunction with other imaging techniques in order to identify correlations between changes in the distribution of important chemical species and other changes in the properties of the tissue. Here we review the applications of mass spectrometry imaging in neuroscience research and discuss its potential. The results presented demonstrate that mass spectrometry imaging is a useful experimental method with diverse applications in neuroscience.
Similar content being viewed by others
INTRODUCTION
Biochemical processes, including those that take place in the brain, revolve around the interactions between various molecules and molecular complexes, and their dynamic spatial redistribution. To understand these complicated biochemical interactions, it is necessary to identify the molecular entities involved and to determine their spatial distribution. Ideally, then, the analytical tools that are used to study these processes should provide as much information as possible about the identities and topographical distributions of the chemical species present within biological samples. Various analytical technologies have been developed for this purpose and several molecular tools have found widespread use for visualizing and identifying molecular targets in the brain, including chemical stains, DNA probes, immunohistochemical tags, and radiolabeled ligands. Analytical technologies that rely on these tools can generate multidimensional images with very high temporal and spatial resolutions. However, most of them can only be used with prior knowledge about the chemical species of interest. In addition, they generally require that the target is preselected, by labeling or with affinity tags or expression markers, before analysis. This limits the amount of chemical information that can be obtained.
Over the last two decades, mass spectrometry (MS) has become an important analytical technique in neuroscience research. The introduction of two soft ionization techniques, matrix-assisted laser desorption ionization (MALDI), and electrospray ionization (ESI), has made it possible to detect and quantify proteins, peptides, lipids, and metabolites, which has proven particularly useful in large-scale ‘-omics’ profiling studies. Investigations of this type, including expression profiling studies and those using functional approaches have become much more common in recent years. Due to the widely divergent molecular compositions and physicochemical properties of proteins, peptides, and lipids, a range of techniques have been introduced for use in these investigations, most of which depend on the integration of biological, chemical, and analytical methods. MS-based ‘omics’ technologies and sophisticated bioinformatics tools have provided numerous unexpected insights into the functioning of diverse biological systems (Bayes and Grant, 2009) and have great potential for the discovery of novel biologically relevant molecules (Svensson et al, 2007; Svensson et al, 2003). However, their usefulness in neuroscience research may be limited due to their inability to provide information on the spatial distribution of specific analytes within brain tissue samples.
MS imaging is a technique that can be used to investigate the spatial distribution and topographical organization of numerous different molecular analytes on complex surfaces such as brain tissue sections. In mass spectrometry, the analytes are ionized and the mass-to-charge (m/z) ratio of the resulting ions is determined, giving an indication of each analyte’s atomic composition. Hundreds of molecules can be detected and identified simultaneously at μm or even sub-μm spatial resolutions in complex biological samples. In addition, it is not generally necessary to label the analytes before analysis, which makes it possible to detect and image both known and unknown targets without having to develop and prepare specialized labels (which can be challenging in and of itself). The first MS imaging experiments were performed 60 years ago, using secondary ion mass spectrometry (SIMS) to image elemental ions (Castaing and Slodzian, 1962; Liebl, 1967). The first molecular imaging experiments were performed in the 1970s, using SIMS and laser microprobe mass analysis (Hillenkamp et al, 1975). SIMS is a surface analysis technique that works by exposing the tissue sample’s surface to a focused primary ion beam and then collecting and analyzing the resulting secondary ions. A wide range of ion sources and mass analyzers can be used in SIMS experiments, but time-of-flight (TOF) analyzers and cluster ion sources are the most popular tools for studying biomolecules in this way.
Subsequent to the development of SIMS, a number of other mass spectrometric techniques have been introduced and used in imaging studies with biological samples. These methods differ primarily with respect to the method of ionization; the most notable are desorption ESI mass spectrometry (DESI-MS), inductively coupled plasma mass spectrometry (ICP-MS), and MALDI-MS.
DESI is an atmospheric pressure ionization technique in which an electrospray, an electrically charged mist of an appropriate solvent, is directed to the sample surface to ionize chemical species. Ionization occurs via the interaction of the charged microdroplets from the spray with the chemical species in the surface of the sample (Takats et al, 2004). A modified version of the technique known as DESI-MS imaging has been developed that can be used to generate two-dimensional images showing the distribution and concentration of analytes in a tissue section (Ifa et al, 2007)
In ICP-MS, the sample is ionized by exposure to a plasma generated by electromagnetic induction. This method can be used to analyze metals and some non-metals in samples with high sensitivity. Laser ablation (LA) ICP-MS, which uses a laser beam to ablate material from the surface to facilitate ionization, has shown considerable potential in the imaging of biological tissues. It can be used to determine the localization of many elements in biological sections with high sensitivity and relatively high spatial resolution (Becker et al, 2005; Qin et al, 2011).
The use of MALDI-MS for the imaging of biological tissue samples was first described by Caprioli et al (1997). MALDI-MS imaging of brain tissue samples is usually performed in situ with thin (10 μm) tissue sections. The sections are typically coated with a uniform layer of the matrix crystals, after which an ordered array of mass spectra is acquired from raster-defined grid points. Commonly used MALDI matrices are sinapinic acid, α-cyano-4-hydroxycinnamic acid, and 2,5-dihydroxybenzoic acid (DHB). Consequently, each mass spectrum reflects the local molecular composition at a given pair of x and y coordinates. Images showing the distribution of selected molecules within the tissue section are then generated by extracting data for the corresponding m/z ranges from the spatially acquired MS data files (Fig. 1). This provides information on the spatial localization of the molecules within the sample without needing extensive sample preparation (Norris and Caprioli, 2013; Stoeckli et al, 2001). All of the MS imaging techniques discussed in this section differ in terms of their specificity, sensitivity, and resolution and are therefore best suited to different classes of analytical problems.
General overview of matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) imaging. Tissue sections from frozen brain are cut by a cryomicrotome and transferred to a conductive glass slide. Tissue sections are coated with a specific matrix, depending on the nature of the biomolecule to be analyzed. MALDI-MS imaging allows simultaneous mapping of hundreds of multimodal substances in thin tissue sections with a lateral resolution down to a few μm. From a raster over the tissue and measurement of the peak intensities over thousands of spots, mass spectrometric images are generated at specific molecular weight values.
MASS SPECTROMETRY IMAGING OF MULTIMODAL MOLECULES
The brain contains numerous endogenous compounds that are involved in signaling, biosynthesis, and metabolic processes. These compounds have a wide range of chemical identities: some are simple alkali metal ions, while others are large proteins and multi-protein complexes. The spatial distributions of these compounds, categorized below, are as important as their chemical properties, because both their localization and their concentration change in many neurodegenerative diseases and during specific neurological events. The most popular MS imaging techniques for ex vivo mapping of the distribution of the compound classes discussed below are MALDI, DESI, TOF-SIMS, and ICP.
In general, sample preparation methods for MS imaging should maintain the localization of the analyte of interest without causing any degradation. In typical protocols, the organs of interest are rapidly removed by dissection and snap-frozen in a liquid nitrogen-isopentane bath (Jehl et al, 1981). It has been shown that post-mortem degradation can occur very quickly, producing false-positive and/or negative results in subsequent imaging studies (Goodwin et al, 2008; Goodwin et al, 2012; Goodwin et al, 2010; Skold et al, 2007).
One way of suppressing degradation reactions in isolated tissue samples involves the use of thermal irradiation (Svensson et al, 2009); formaldehyde-fixed paraffin-embedding should be avoided because it causes protein-peptide crosslinking. Tissue sections are prepared by cutting the organs into slices with a thickness of less than 20 μm. The use of supporting media such as optimal cutting temperature (OCT) compound should not be used. Tissue sections are either attached to stainless steel plates or indium-tin-oxide glass slides for analysis using high vacuum MALDI instruments, or to non-conductive glass slides for intermediate pressure instruments. Cell debris, lipids, salts, and other signal-suppressing components of the tissue that interfere with the ionization/desorption of proteins and peptides may be selectively removed by washing the section in a way that fixes the tissue sample and preserves the localization of the target compounds. Various solvents and treatment strategies have been used for this purpose (Lemaire et al, 2006; Seeley et al, 2008), but the protocol that we have found to be most useful in preparing samples for MALDI-MS imaging of neuropeptides involves washing the tissue with 70% ethanol for 10 s and then with 95% ethanol twice for 10 s each time.
Matrix application is an important step that has a significant effect on the quality of the results obtained in the imaging experiment. The matrix can either be deposited as a uniform layer (using automatic or manual pneumatic sprayers, or by sublimation in the case of dry matrices) or can be applied as a series of micro spots (using piezo- or acoustic spotters). Wetter matrix application conditions may be preferred as they facilitate the extraction of proteins and peptides. However, they tend to produce relatively large matrix crystals, which limit the spatial resolution of the resulting images. Once the matrix has been applied, the prepared sample is loaded into an imaging-capable MALDI instrument and spectra are acquired for the predefined region of interest and raster size.
MS imaging of peptides and proteins
Of the existing mass spectrometry imaging techniques, MALDI-MS imaging has proved to be the most useful for determining the localization of proteins and peptides (Chaurand et al, 2002). DESI-MS can also be used to study the distribution of peptides (Pan et al, 2013) and proteins (Shiea et al, 2008), as well as multi-protein complexes (Ferguson et al, 2011; Przybylski et al, 2010) in concentrated spotted samples, but will require further refinement in order to detect proteins and peptides directly in tissue samples. Despite the high spatial resolution achieved using TOF-SIMS imaging, it has only limited applications in the imaging of proteins and peptides because its mechanism of ionization causes extensive fragmentation of the target compounds, which makes their identification difficult. However, it has been used to map the localization of the protein thyroglobulin in pig thyroid glands after trypsin digestion by comparing the small peptide fragments produced from tissue samples to those produced by analyzing standard solutions of the target proteins (Nygren and Malmberg, 2010). The use of a softer ionization method based on a Bi-cluster ion source makes it possible to analyze peptides with masses of up to 2 kDa by TOF-SIMS imaging (Komatsu et al, 2008). MALDI-MS imaging produces results with a lower spatial resolution than TOF-SIMS, and requires that the matrix be deposited on the tissue. However, it uses much milder ionization conditions and has thus been more widely used for protein and peptide imaging.
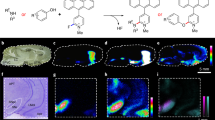
The advantages of MALDI-MS imaging have made it the most popular MS technique for the imaging of proteins (Burnum et al, 2008; Caprioli et al, 1997; Chaurand et al, 2002; MacAleese et al, 2009; Pierson et al, 2004; Skold et al, 2006). We recently presented a novel MALDI-MS imaging protocol for the quantitation of an endogenous neuropeptide directly in tissue sections. Substance P 1–11 (SP) was quantified in different mouse brain anatomical structures. The SP levels correlated well with previously reported peptide levels using other techniques. To confirm the identity of SP, tandem MS (MS/MS) was performed directly on tissue sections and the obtained MS/MS spectra from tissue sections were compared with spectra obtained from synthetic SP (Kallback et al, 2012) (Figure 2).
Quantitation of endogenous substance P 1–11 (SP) concentration in a mouse brain sagittal tissue section. (a) Digital photograph of the mouse brain tissue section indicating different brain tissue structures. (b) SP was quantified in the nucleus accumbens, caudoputamen, globus pallidus external segment, substantia nigra, substantia innominata. The matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) imaging analysis shows that endogenous SP is detected at high concentrations in substantia innominata, globus pallidus external segment, and substantia nigra. (c) Mass spectrum from the substantia innominata structure showing the monoisotopic mass of SP (inset). Reproduced with permission (Kallback et al, 2012).
However, the in situ identification of unknown compounds by MALDI-MS remains challenging. Both bottom-up and top-down proteomics identification approaches for the in situ identification of proteins and peptides have been investigated. In the bottom-up approaches, the proteins are digested in situ using a protease enzyme (typically, trypsin) and the resulting peptides are analyzed either by MS/MS fragmentation or by peptide mass fingerprinting (Groseclose et al, 2007; Stauber et al, 2010). The main problems encountered when using this approach relate to the complexity of the digested sample and the risk of delocalization during the digestion reaction(s). It has been demonstrated that the quality of the peptide identification process and the number of successfully identified peptides can be increased by performing a liquid chromatography (LC)-ESI MS/MS analysis of an adjacent tissue section in parallel with in situ bottom-up identification experiments when working with mouse brain samples (Schober et al, 2011). In the top-down identification approaches, the intact molecular ions of the proteins or peptides generated by a soft ionization technique are subjected to gas phase fragmentation in the mass spectrometer. The resulted product ions are used for identification of the parent protein or peptide.
In-source decay (ISD) is the most widely used activation technique in MALDI-MS experiments and can be used to fragment protein and peptide ions in order to obtain sequence data when using the top-down approach (Bonnel et al, 2011; Debois et al, 2010). However, the efficiency of this method is necessarily determined by the tendency of the target protein or peptide to fragment under ISD conditions. Recently, the high mass accuracy and resolution of a MALDI-Fourier transform ion cyclotron resonance instrument (FT-ICR) MS were exploited to perform targeted in situ identification of proteins using ISD. By comparing the ISD fragments observed when analyzing samples in imaging mode to reference spots, these authors were able to identify target proteins such as myelin basic protein in mouse brain sections (Calligaris et al, 2013).
MS imaging of lipids
Lipids have essential roles in numerous cellular processes and are involved in many diseases, including neurodegenerative disorders (Puglielli et al, 2003). Because of their relatively high abundance in biological samples, they were one of the first compound classes to be examined in MS imaging studies. TOF-SIMS, MALDI, and DESI-MS imaging are all widely used to map the localization of lipids. TOF-SIMS has proven to be capable of mapping the localization of diverse lipids, including glycerophospholipids (Sjovall et al, 2006; Sjovall et al, 2004), sterol lipids (Borner et al, 2006; Sjovall et al, 2004), and sphingolipids (Borner et al, 2006; Nygren et al, 2005; Pernber et al, 2007; Sjovall et al, 2006; Sjovall et al, 2004) with sub-micron spatial resolution. However, its utility is limited by its tendency to cause extensive fragmentation of the target analytes and its comparatively low sensitivity in terms of secondary ion yields (Sjovall et al, 2004). Some of these difficulties can be partially overcome by using cluster ion sources based on Au3+.
In a notable recent study, TOF-SIMS imaging was used to investigate lipid distributions within mouse brain sections. The samples were prepared by simple cryosectioning, then rinsed in ammonium formate, and freeze dried before undergoing TOF-SIMS analysis. Cholesterol, sulfatides, and phosphatidylcholines were all identified; the images obtained showed that the gray matter is rich in phosphatidylinositols, phosphatidylcholines, and sulfatides, while the spectra for the white matter were dominated by cholesterol-related peaks (Sjovall et al, 2004). In a subsequent investigation, cresyl violet and eosin Y staining were used to obtain complementary information on the structures observed in the TOF-SIMS images of freeze-dried mouse brain sections (Sjovall et al, 2006). Further structural information was obtained by TOF-SIMS imaging of rat cerebellar white matter (Borner et al, 2006) and cortex samples (Nygren et al, 2005) using a Bi+ ion cluster source. These investigations demonstrated that the cerebellar white matter contains previously unknown cholesterol-rich ribbon-shaped areas that are 10–20 μm long in which Na+ and K+ are excluded, and that galactosylceramide (GalC) C18 is predominantly located in cholesterol-rich areas, whereas GalC C24 is localized in regions that are rich in Na+ and K+ (Borner et al, 2006).
To investigate the scope for depth profiling/imaging of biological tissues such as rat brain samples, a fullerene ion beam was used to sputter the surface of a rat brain tissue section. Although the damage induced by the fullerene sputter gun was much less severe than that caused by heavy metallic ion beams, it was still sufficient to make TOF-SIMS depth profiling very challenging (Debois et al, 2007).
Because of the remarkable spatial resolution of TOF-SIMS, it is one of the few MS techniques that can be used to visualize and identify individual lipids at the cellular and subcellular levels in cells such as neurons (Passarelli and Winograd, 2011; Sjovall et al, 2003; Yang et al, 2010a; Yang et al, 2010b). However, there is a trade-off between high resolution and sensitivity, which limits the utility of this technique for studying less abundant compounds at the (sub)cellular level.
The first application of DESI-MS in imaging involved the visualization of lipids in rat brain tissue sections (Ifa et al, 2007). Depending on the solvents used and the nature of the substrate, spatial resolutions of around 200 μm could be achieved. However, it has been reported that imaging with a spatial resolution of around 12 μm is possible with a nano-DESI instrument (Laskin et al, 2012). The mass resolution of DESI-MS imaging was improved substantially by combining the apparatus with a FT-ICR mass spectrometer. This made it possible to resolve signals from mouse brain lipids that overlap when analyzed using more conventional instruments (Pol et al, 2009). DESI-MS imaging has also been used to analyze and characterize the lipid profiles of different human astrocytoma subtypes, showing that some marker lipids have different abundances in different subtypes (Eberlin et al, 2010a). Most studies that have used DESI-MS for the imaging of brain lipids have examined experimental models and were conducted for the purposes of method development (Eberlin et al, 2010b; Laskin et al, 2012; Pol et al, 2009; Wiseman et al, 2006) or to evaluate sample preparation protocols (Dill et al, 2011; Laskin et al, 2012) rather than to answer neurological questions.
MALDI-MS imaging has been used extensively to map the distribution of lipids in a wide range of organs, including the brain. Benabdellah et al (2010) compared MALDI-MS and TOF-SIMS imaging for the visualization of rat brain lipids. It was concluded that MALDI-MS imaging is a robust and reproducible technique provided that care is taken during sample preparation and matrix application. The two techniques were found to have different advantages: MALDI-MS imaging was capable of nM sensitivity, whereas TOF-SIMS was able to achieve sub-μm spatial resolutions (Benabdellah et al, 2010). To map the distribution of brain lipids many MALDI-MS imaging studies were conducted for the purposes of method development; notably, several have focused on the identification of suitable matrices and salts for use with this technique (Ageta et al, 2009; Astigarraga et al, 2008; Cerruti et al, 2012; Cerruti et al, 2011; Chan et al, 2009; Chen et al, 2008; Colsch and Woods, 2010; Franck et al, 2009; Goto-Inoue et al, 2010; Meriaux et al, 2010; Puolitaival et al, 2008; Shanta et al, 2011; Shrivas et al, 2010; Sugiura and Setou, 2009b; Thomas et al, 2012; Wang et al, 2008; Yang and Caprioli, 2013). Other studies have focused on sample preparation techniques (Angel et al, 2012; Carter et al, 2011; Delvolve and Woods, 2011; Hankin et al, 2007; McLean et al, 2007; Trimpin et al, 2010), improving the capabilities of MALDI-MS imaging instruments (Shimma et al, 2008; Trim et al, 2010), and data processing (Sugiura et al, 2009a; Trim et al, 2008). These studies have also provided spatial information on the localization of many lipids in brain tissue sections.
MS Imaging of neurotransmitters
It can be difficult to use MS techniques such as TOF-SIMS, DESI, and MALDI-MS imaging to study the distribution of small molecules, especially in lipid-rich samples such as brain tissue sections. In TOF-SIMS experiments, the extensive fragmentation of large molecules makes the small molecule region of the spectrum very complex and so it is difficult to identify signals arising from small molecules. This problem has been partially overcome by the development of cluster ion sources (Touboul et al, 2004). Notably, TOF-SIMS with a bismuth-based cluster ion source has been used to visualize the distribution of lipids and some abundant small molecules (including cholesterol) in rat brain sections with a spatial resolution of around 400 nm (Touboul et al, 2004).
Catecholamines (including epinephrine and norepinephrine) have been imaged by DESI-MS in porcine and rabbit adrenal glands with a spatial resolution of 200 μm. The results obtained confirmed previous reports that the levels of epinephrine and norepinephrine in rabbits’ adrenal glands are lower than those in porcine adrenal glands (Wu et al, 2010). DESI-MS has also been successfully used to image cholesterol in adrenal gland and mouse brain samples after reactive derivatization by treatment with betaine aldehyde (Wu et al, 2009).
As DESI-MS is mechanistically very similar to ESI, ion suppression effects caused by abundant and readily ionized compounds such as lipids may interfere with the detection of compounds of interest. This problem of low sensitivity could conceivably be overcome by derivatizing the analytes of interest, by converting them into adducts or by changing the polarity (Manicke et al, 2008).
There are a number of significant technical difficulties associated with MALDI-MS imaging of neurotransmitters, and considerable effort has been invested into finding ways of addressing them. The use of MALDI-MS (and thus MALDI-MS imaging of small molecules) has historically been hindered by the formation of matrix fragments and cluster peaks, which can mask the signal of interest. The recent introduction of a deuterated form of α-cyano-4-hydroxycinnamic acid, the most common matrix used in studies of small molecules, resulted in the unmasking of previously hidden compounds such as acetylcholine. This approach was used to map the localization of acetylcholine and some other endogenous compounds in rat brain tissue sections; acetylcholine was found to be highly abundant in the cerebral cortex, corpus callosum, ventral hippocampal commissure, thalamus, and cerebellum (Shariatgorji et al, 2012b) (Figure 3). In another study, a known MS/MS transition of acetylcholine was examined to enhance the sensitivity of the system in order to visualize the localization of acetylcholine in mouse brain sections (Sugiura et al, 2012). Surface-assisted laser desorption ionization MS replaces the chemical matrix used in MALDI with inorganic surfaces and nanomaterials, thereby providing an alternative way of overcoming the matrix peak interference problem in MALDI-MS (Shariatgorji et al, 2009; Wei et al, 1999). Titanium dioxide nanoparticles have been used successfully for this purpose in a study that mapped the distribution of endogenous low molecular weight compounds such as gamma aminobutyric acid in mouse brain tissue sections (Shrivas et al, 2011).
Matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) imaging relative abundance and spatial distribution of the neurotransmitter acetylcholine (m/z 146.1±0.1) in a rat brain sagittal tissue sections. The confirmation that m/z 146.1 is acetylcholine was achieved by on tissue MS/MS fragmentation, where product ions of m/z 87.0 and m/z 104.1 were generated and agreed with a MS/MS spectrum from synthetic acetylcholine. The MALDI-MS image shows that acetylcholine is most abundant in brain structures such as cerebral cortex, corpus callosum, ventral hippocampal commissure, thalamus, and cerebellum. Reproduced with permission (Shariatgorji et al, 2012b).
Some neurotransmitters are present only at low levels in brain samples and/or are difficult to ionize. It will therefore be necessary to develop methods with better signal-to-noise ratios than can be achieved with existing technologies in order to map their distribution in biological samples by MS imaging. Methods that could potentially be used to this end include the use of optimized matrices (Cohen and Gusev, 2002), chemical derivatization (Chacon et al, 2011), and selective tissue clean-up (Shariatgorji et al, 2012a).
MS imaging of pharmaceuticals
The imaging of pharmaceuticals and their metabolites in their target sites is a very important source of data in drug discovery and development that can provide information on pharmacokinetics, toxicology, and ADME (absorption, distribution, metabolism, excretion). Despite the difficulties of using MS imaging to analyze small molecules described in the previous section, TOF-SIMS, DESI-MS, and MALDI-MS imaging have found applications in mapping the distribution of pharmaceuticals. However, the latter two techniques have been more popular because of their greater sensitivity and softer ionization mechanisms compared with TOF-SIMS.
Other molecular imaging techniques, including in vitro autoradiography and in situ hybridization histochemistry, have contributed significantly to the visualization of biological processes and have proven to be useful in drug discovery. However, these imaging methods require the molecular labeling of the target compound and it is the distribution of the label that is detected and mapped, even when the original compound has been metabolized or otherwise modified. In the last decade, the pharmaceutical community has expressed considerable interest in MS imaging because unlike most other imaging techniques, MS detects the target molecules directly based on their characteristic m/z ratios and does not require the use of labeled compounds. It can thus potentially be used to detect and characterize drugs, drug metabolites (and endogenous drug-modified molecules) in their native states in brain tissue sections.
The atypical antipsychotic clozapine was imaged using MALDI-MS, which had sufficient spatial resolution and sensitivity to reveal the drug’s distribution in rat brain tissue samples. The images of clozapine in rat brain sections obtained using the MALDI-MS method were consistent with those obtained by autoradiography (Hsieh et al, 2006). The ambient ionization method DESI-MS has been used to perform direct, high-throughput imaging of clozapine in order to directly determine the drug’s distribution within histological brain sections (Wiseman et al, 2008). By scanning the tissue sections with the DESI spray, detailed spatial maps were generated for many molecules simultaneously, providing information on their molecular weights, abundance, and distribution in space. In addition, quantitative data on the abundance of clozapine in brain samples (and also lung, kidney, and testis samples) obtained using DESI were compared with those obtained by conventional LC-MS/MS methods. The two data sets were in good agreement, which suggests that DESI imaging may be useful for the direct and quantitative detection of drugs and drug metabolites in biological tissue samples (Wiseman et al, 2008).
In addition to its applications in mapping the distribution of pharmaceutical agents, we have shown that MALDI-MS imaging can be used to directly and quantitatively determine the absolute concentrations of target compounds in specific regions of tissue sections (Goodwin et al, 2011; Nilsson et al, 2010). This was done by comparing the signal intensities for the regions of interest in dosed tissue samples to those for quantitation control spots adjacent to the regions of interest. The quantitation control spots were created by spotting a known amount of the target compound directly onto the surface of the sample.
One drug discovery area where MS imaging has found a growing range of applications is the development of novel positron emission tomography ligands for CNS-disorders. It is particularly useful in this context because it can simultaneously provide information on the distribution of compounds within the brain and show how their concentration changes over time. Two widely used reference PET ligands, raclopride and SCH 23390, were given systematically and then directly imaged in post-mortem rat brain samples using MALDI-MS (Goodwin et al, 2011). The compounds were identified by examining the accurate mass data and by performing MS/MS fragmentation experiments, and were quantified by analyzing their signal intensities. The distribution of the compounds was determined as a function of the dose administered, and time courses showing the changes in their distribution were acquired at each dose. In addition, quantitative data were obtained by spotting known amounts of the target compound onto the surfaces of control tissue samples and using these as a source of reference data in the evaluation of signal intensities from other regions of the sample. These results clearly demonstrate that MS imaging can be a powerful tool in PET ligand research and development.
It is essential to take care during the sample preparation step when using MALDI-MS imaging to study small molecules, for the same reason that similar care is essential when studying proteins and peptides. It has been shown that the detected abundance of drug molecules in brain tissue samples decreases with the length of time they spend at room temperature. This might be because proteolysis increases the abundance of peptides in the sample over time, or because of direct modification or metabolism of the drug molecules themselves (Goodwin et al, 2012).
Fluctuations in the intensity of MALDI-MS imaging signals sometimes arise from inhomogeneity in the matrix coating, structural differences within the sampled tissue, or laser instability. These can be compensated by normalization against the total ion count, root mean square or median signal intensity, or by using labeled compounds as internal standards. We have recently shown that labeled normalization using a deuterated analog of the target compound is more effective than alternative methods of compensating for false ion signals. Based on this finding, deuterated imipramine was used as a normalization reference compound in order to quantitate the levels of imipramine in mouse brain tissue sections (Kallback et al, 2012). In another study, cocaine was quantified in brain tissue samples using deuterated cocaine as an internal standard. Scan-by-scan normalization using wide-isolation MS/MS improved the linearity of the calibration curve as well as the signal reproducibility, making it possible to obtain robust quantitation results that were comparable to those obtained using LC-MS/MS (Pirman et al, 2013).
MS Imaging of inorganic ions
Metal ions have important roles in many signaling and metabolic pathways due to their diverse redox properties and varied coordination chemistry. Potassium, calcium, and sodium have functions in signal transduction, synaptic transmission, plasticity and cell excitability. Transition metals such as zinc (Sensi et al, 2011), copper (Monnot et al, 2011), manganese (Sloot and Gramsbergen, 1994), and iron (Zecca et al, 2004) have essential roles in neurotransmitter synthesis, the regulation of synaptic transmission, and brain development. TOF-SIMS is a highly sensitive technique for the imaging of inorganic ions in biological tissues with a high spatial resolution. It has been used to image sodium, potassium, magnesium, and calcium ions in retinal tissues (Kim et al, 2008) and aluminum in human brain sections (Candy et al, 1992). In LA-ICP-MS, a laser beam is used to ablate material from solid surfaces before mass spectrometric analysis. There has been some interest in using this method for elemental imaging of biological samples as it provides more reliable quantitative data than TOF-SIMS, albeit with a lower spatial resolution. It is also a very sensitive technique (with sub-μg/g limits of detection) and can be used to perform elemental imaging of ions that are present at trace concentrations in thin tissue sections. It has therefore been used widely for elemental analysis of brain samples (Becker et al, 2008; Becker et al, 2007; Becker et al, 2005; Dobrowolska et al, 2008; Qin et al, 2011; Zoriy and Becker, 2007; Zoriy et al, 2008; Zoriy et al, 2006). Notably, it was used to quantitatively analyze the distributions of multiple metallic elements in 20 μm thick human brain sections with a spatial resolution of 50 μm. Matrix-matched standards were used to quantitate zinc, uranium, copper, and thorium in human hippocampus samples, revealing that the distributions of phosphorous, sulfur, copper, and zinc were inhomogeneous, whereas uranium and thorium were present at lower concentrations but were relatively homogeneously distributed (Becker et al, 2005). In another effort, zinc, copper, and lead were imaged and quantified in the insular, central, and hippocampal areas of the human brain. The highest concentrations of zinc and copper occurred in the hippocampus, while lead had a more homogenous distribution (Dobrowolska et al, 2008). More recently, the possibility of using MALDI-MS and LDI-MS imaging to examine the distribution of metallic elements in brain tissue sections has been explored, with promising results. We have recently obtained MALDI-MS images using this approach, which was used to study the distribution of potassium ions in sagittal sections of rat brains at a spatial resolution of 100 μm using a DHB matrix (Figure 4).
(a) Optical image and (b) matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) imaging relative abundance and distribution of potassium (m/z 39) of a rat brain coronal section. Dashed line shows the measured region and data are displayed in rainbow scale over the same range (scale bar, 5 mm). (c) The overall mass spectrum of the rat brain coronal section shows the signal of potassium and sodium ions.
MS imaging of endogenous metabolites
Metabolites, including metabolic intermediates such as lipids, amino acids, organic acids, and small signaling molecules, are the end products of cellular regulatory processes. Cellular metabolic processes utilize adenosine triphosphate (ATP) as an energy source, converting it into adenosine diphosphate (ADP) or adenosine monophosphate (AMP). A MALDI-MS imaging approach was developed for the simultaneous visualization of these adenosine nucleotides in order to provide information on energy production and consumption in brain tissue sections. The method proved to be capable of directly detecting and identifying 13 primary metabolites in rat brain sections, at a spatial resolution of 50 μm (Figure 5): AMP, ADP, ATP, uridine monophosphate, cyclic AMP, fructose-1,6-bisphosphate, guanosine monophosphate, uridine diphosphate (UDP), guanosine diphosphate, guanosine triphosphate, ADP-ribose, UDP-glucose, and UDP-N-acetylglucosamine (UDP-GlcNAc) (Benabdellah et al, 2009). These experiments opened a new field of investigation for MS imaging of primary metabolites in situ.
Matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) images of (a) adenosine monophosphate (AMP), (b) adenosine diphosphate (ADP), (c) UDP-N-acetylglucosamine (UDP-GlcNac), (d) fructose-1,6-bisphosphate (F-1,6-biP), and (e) guanosine triphosphate (GTP) acquired in the negative ion mode from a rat brain section deposited on a stainless steel plate, after deposition of a homogeneous layer of 9-AA. Field of view: 8.3 × 8.3 mm2, 50 μm spatial resolution. The values of intensity (I) indicated under each image correspond to the minimal and maximal intensity in a pixel. (f) Optical image of a brain tissue section after 9-aminoacridine deposition as matrix and analysis by MALDI-MS imaging with a 50 μm spatial resolution. Reproduced with permission (Benabdellah et al, 2009).
MASS SPECTROMETRY IMAGING IN NEURODEGENERATIVE DISEASES AND NEUROPSYCHOPHARMACOLOGY
Alzheimer’s disease
Alzheimer’s disease (AD) is the most common form of dementia. With an aging population, its frequency is increasing rapidly around the world. AD is characterized by protein misfolding in the brain and this process and the associated brain damage begin years before the onset of dementia (Perrin et al, 2009). Loss of neurons and synapses occurs in the cerebral cortex, hippocampus, and certain subcortical regions, which results in gross atrophy of the affected regions (Wenk, 2003).
It has been suggested that the pathological sequence of events in AD is caused by plaques in the brain (Hardy and Allsop, 1991). According to this hypothesis, the accumulation of amyloid-beta peptides (Aβ) is the primary factor that drives AD pathogenesis. Other processes of the disease, including the formation of neurofibrillary tangles containing tau protein, interact with Aβ production, and Aβ clearance (Hardy and Selkoe, 2002).
Efforts are gradually being focused on finding alternative theories to the Aβ cascade hypothesis and treatments that do not involve removing Aβ plaques. The apolipoprotein epsilon 4 variant is the largest known genetic risk factor for late-onset sporadic AD in a variety of ethnic groups and there is a growing body of evidence that supports the involvement of lipids in the pathogenesis of AD (Hirsch-Reinshagen et al, 2009; Puglielli et al, 2003). MS imaging can be used to obtain detailed pictures of the distribution of compounds associated with the Aβ cascade, lipids, and neuropathology in human and animal brain tissue samples simultaneously, thereby potentially providing information that could be used to further test and compare the competing hypotheses.
The laminar distribution of cholesterol in the cerebral cortex of brain samples from control subjects and AD patients has been investigated using TOF-SIMS imaging (Lazar et al, 2013). It was shown that cholesterol was unevenly distributed along the cortical thickness, being more abundant close to the white matter in both the control and AD samples. However, the AD samples had 34% more cholesterol in the gray matter of the cerebral cortex than did the controls, and were observed to have accumulations of cholesterol in the lower half of the cortex (cortical layers III and IV). This was the first time that MS imaging had been used to visualize the distribution of cholesterol in the human cortex and the first attempt to specifically identify potentially pathological changes in its distribution within the cortical layers. It was suggested that the enrichment of cholesterol in neuronal membranes might promote the endocytosis of amyloid precursor protein and thus its cleavage, leading to the production and aggregation of Aβ peptide (Chochina et al, 2001; Cossec et al, 2010). If this was the case, the accumulation of cholesterol in the brain might be an attractive therapeutic target.
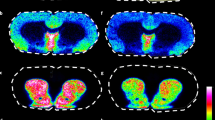
MALDI-MS imaging has been used to perform a spatial analysis of the distribution of various Aβ peptides in an animal model of AD (transgenic mice expressing human APP751 with the Swedish mutation) (Rohner et al, 2005; Stoeckli et al, 2002). This model has a mutation in the gene that encodes the amyloid precursor protein, which causes increased production of Aβ and Aβ-peptide fragments and results in the development of significant Aβ deposits in the cortical and hippocampal structures as the animals are aging. MALDI-MS imaging demonstrated that Aβ peptides 1–40 and 1–42 were the most abundant amyloid peptides in these areas of the brain. Furthermore, experiments on the mapping of multiple Aβ peptides in the APP23 transgenic mouse AD model showed that Aβ peptides 1–37, 1–38, 1–39, 1–40, and 1–42 could be imaged simultaneously (Stoeckli et al, 2006) (Figure 6). Clustering and multivariate analysis were used to identify and visualize spatial and spectral correlations in the MS images. Similarly, TOF-SIMS imaging has been used to demonstrate that cholesterol is selectively localized near inclusion bodies in transgenic mouse AD models (Sole-Domenech et al, 2013) (Figure 7). This is consistent with the results of previous studies on the interactions between transmembrane cholesterol and amyloid precursor proteins, which suggested that this interaction is important in Aβ formation and AD pathology (Barrett et al, 2012).
Matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) images of amyloid-beta peptide (Aβ) peptide distributions in mouse brain tissue section from Alzheimer’s disease (AD) mouse model (APP23). Within one image acquisition, multiple Aβ peptide are measured simultaneously (individual mass peaks are used to generate separate images). Reproduced with permission (Stoeckli et al, 2006).
Cholesterol accumulates in the vicinity of amyloid-beta peptide (Aβ) deposits in 3 × Tg-Alzheimer’s disease (AD) mouse model brain tissue sections. Images of two areas located in the superior subiculum show the time-of-flight secondary ion mass spectrometry (TOF-SIMS) cholesterol distribution (a–c) and the overlaid cholesterol/p-FTAA images (b–d) (p-FTAA is an amyloidotropic fluorescent dye that binds primarily to fibrillary Aβ). Cholesterol accumulations are highlighted by white arrowheads in (a–c). Reproduced with permission (Sole-Domenech et al, 2013)
Sulfatides have been imaged in cerebral cortex tissue sections of the human brain using MALDI-MS imaging, which revealed that the relative abundance of non-hydroxylated and hydroxylated sulfatides is reversed on crossing the boundary that separates the white and the gray matter (Yuki et al, 2011). This study also showed that the sulfatide levels in cerebral cortex samples from AD patients were identical to those in control samples, whereas previous reports suggested that there was a significant difference in this respect (Han et al, 2002).
The results discussed above clearly show that MS imaging can provide valuable molecular information on the content and distribution of Aβ peptides, lipids, and lipid precursors in pathological and non-pathological tissues. As TOF-SIMS is comparatively non-destructive, tissue samples analyzed using this method can subsequently be studied using complementary techniques (Sole-Domenech et al, 2013). This makes it possible to investigate the role of the various potential biological components of AD in multiple ways, which would be virtually impossible if one were to use only one single method of imaging and analysis.
Parkinson’s disease
Parkinson’s disease (PD) is a slowly progressive neurodegenerative disorder that affects 1% of the population over the age of 60. Its typical symptoms include slowness of movement, rigidity, resting tremors, and postural instability (Lees et al, 2009). Histologically, PD is characterized by a loss of dopamine neurons in the substantia nigra, which reduces dopamine signaling in the nigrostriatal pathway. The progression of PD correlates with the abundance of dopamine (and its metabolites) and also with the expression of certain peptides in the striatal region. For example, it is well known that the regulation of opioid peptide synthesis in the basal ganglia is disturbed in PD (Bezard et al, 2001).
MALDI-MS imaging has been used to study changes in the abundance of different molecular species in PD animal models. In an early attempt to identify phenotypic markers of PD, a histological approach was used to study protein expression in an experimental model in which PD-like symptoms were induced by the unilateral administration of 6-hydroxydopamine (6-OHDA) (Pierson et al, 2004). Coronal brain sections were then taken and MALDI-MS profiling was used to identify differences in protein expression between the dopamine-denervated and normal hemispheres. The expression profiles revealed several statistically significant changes in the abundance of various proteins (including calmodulin, cytochrome c, and cytochrome c oxidase) in the dopamine-depleted striatum as well as an increase in protein acetylation. Subchronic administration of L-DOPA made these changes less pronounced, which is consistent with the hypothesis that they are disease-specific (Pierson et al, 2004).
Analyses of several different genetic defects have suggested that there is a relationship between the impairment of the ubiquitin conjugation pathways and various forms of PD. Elevated levels of unconjugated ubiquitin were observed in the dorsal striatum of the dopamine-depleted hemisphere but not in the unlesioned hemisphere or the ventral striatum of the dopamine-depleted hemisphere (Pierson et al, 2005). This suggests that the denervation of the dopamine neurons directly affects the regulation of the ubiquitination pathways.
Furthermore, FKBP-12, a 12-kDa FK506-binding protein (neuroimmunophilin), which acts as a receptor for the immunosuppressant drug FK506, showed increased levels in the dorsal and middle part of the 6-OHDA lesioned striatum. In this study, we utilized a number of analytical approaches, such as two-dimensional gel electrophoresis, in situ hybridization, and western blotting to confirm the findings utilizing MALDI-MS profiling directly on brain tissue sections (Nilsson et al, 2007). These early studies highlighted the potential of MALDI-MS imaging by demonstrating its ability to provide valuable insights into the molecular processes involved in PD, to shed light on the underlying pathophysiology of the disease, and to distinguish between metabolic fragments, conjugated proteins, and proteins that have undergone post-translational modification.
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) is a neurotoxin that is commonly used to induce PD symptoms in animal models. MPTP-lesioned mouse brains were used as a PD model to study changes in the levels of various peptides relative to those observed in control samples using MALDI-MS imaging. PEP-19, a neuronal calmodulin-binding protein, was found to be predominantly localized in the striatal region and was around 30% less abundant in the MPTP-lesioned brains (Skold et al, 2006) (Figure 8). This suggests that deficiencies in calcium homeostasis may be partially responsible for the cytotoxic events that are associated with PD neurodegeneration. MALDI-MS imaging has also been used to characterize the spatial distribution of prodynorphin peptides in PD animal models that were subjected to L-DOPA induced dyskinesia. L-DOPA is the most widely used pharmaceutical agent for treating the symptoms of PD, but it can cause severe long-term side-effects such as abnormal involuntary movements (Lees et al, 2009). The MS imaging data revealed changes in the levels of alpha-neoendorphin and dynorphin B in the dorsal lateral striata of animals with higher levels of dyskinesia. Moreover, elevated levels of those two peptides were observed in the substantia nigra reticulate, which is the major output structure of the direct pathway in rats (Ljungdahl et al, 2011). It was also found that the severity of dyskinesia correlates with the abundance of N-terminal tyrosine-cleaved alpha-neoendorphine in the striatum, which may indicate that the inactivation of dynorphin is somehow involved in the induction of dyskinesia (Hanrieder et al, 2011).
Matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) imaging of brain tissue sections after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) administration. The relative ion density of PEP-19 from one control (a) and one MPTP-treated animal (b) shows that there is a reduction of PEP-19 expression in the striatum. Adapted from Skold et al (2006).
Elemental imaging, and particularly metal imaging, has found applications in PD research. Alpha-synuclein and neuromelanin are both known to be important in PD and both interact with metal ions (Double et al, 2003; Rasia et al, 2005). Alpha-synuclein has at least three copper-binding sites and its aggregation properties change upon copper binding. Neuromelanin may have a neuroprotective role due to its high affinity for iron, which may enable it to prevent damage caused by free radicals and oxidative stress. In addition, the similarity of the symptoms of PD and manganism could be taken to indicate that manganese might be also involved in PD. LA-ICP-MS was used to quantitatively image the distributions of copper, manganese, iron and zinc in 6-OHDA induced PD mouse models. It was shown that lesioning caused bilateral increases in the concentration of iron within the substantia nigra but the amounts of copper, manganese, and zinc were comparable in the lesioned and control animals (Hare et al, 2009). In another study, LA-ICP-MS was used to map the distribution of metals at different time points following treatment with MPTP to induce PD-like symptoms. It was found that this caused reductions in the copper content of the periventricular zone and fascia dentate, which is consistent with the observation that MPTP treatment causes the depletion of ATP and thereby affects the availability of energy to drive cellular metabolism. The concentration of copper increased by 40% after 28 days, which is thought to be due to a recovery mechanism. The concentration of iron decreased slightly in the substantia nigra and increased by 40% in the interpeduncular nucleus following MPTP injection. After 28 days, normal iron levels had been restored in the substantia nigra but the iron content of the interpeduncular nucleus remained high (Matusch et al, 2010).
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that involves degeneration of the upper motor neurons of the motor cortex and brainstem and the lower motor neurons of the brainstem and spinal cord together with their associated tracts. The clinical symptoms of upper motor neuron degeneration include spasticity, brisk reflexes, and clonus, whereas the symptoms of lower motor neuron degeneration include weakness, atrophy, hyporeflexia, muscle wasting, and fasciculations (Hardiman et al, 2011).
ALS is currently diagnosed on a clinical basis, and there is generally a significant delay between the onset of symptoms and diagnosis. There is therefore an ongoing search for biomarkers that reflect the progression of the disease and could thus increase the scope for rapid diagnosis as well as providing potential drug targets. Attempts to identify such biomarkers typically revolve around the analysis of blood and cerebrospinal fluid samples from ALS patients, as well as neuroimaging and studies of post-mortem brain tissue.
MALDI-MS imaging has been used to investigate the chemical changes that occur in the proteome of post-mortem spinal cord tissue from ALS patients. It was found that two proteins localized within the cell bodies that contain the central gray matter of the spinal cord were less abundant in the samples from ALS patients than in control samples (Hanrieder et al, 2013). One of these proteins was identified as a truncated version of ubiquitin (Ubc 1–76) that had both of its C-terminal glycine residues removed (Ubc-T/Ubc 1–74), while the other protein remains unidentified (Figure 9). This finding was further validated by western blot and immunohistochemistry experiments. The observation of region-specific ubiquitin processing suggests a disease-related change in protease activity (protein turnover) and thus provides new data on the molecular processes associated with ALS.
Protein intensity differences between amyotrophic lateral sclerosis (ALS) and controls. (a) Protein peaks with m/z 8429 and m/z 8451 were found to be mainly abundant in the gray matter and (b) showed significant lower peak intensity in ALS patients. (c) Unpaired statistical analysis (statistical analysis of microarray data) (SAM) were used (P<0.05). Scale bar, 5 mm. Reproduced with permission (Hanrieder et al (2013).
Stroke
MALDI-MS imaging has been used to characterize the distribution of lipids in an ischemia/reperfusion injury model of the rat brain after bilateral occlusion of the carotid artery. This was found to cause changes in the levels of various phospholipids and ceramides in the hippocampal region; the affected compounds are known to be associated with cellular apoptosis (Hankin et al, 2011). Similarly, Koizumi et al (2010) reported changes in the levels of phosphatidylcholine in the vicinity of a focal cerebral ischemia in a rat model based on MS imaging data. The imaging of brain lipids and gangliosides has been used to study ischemia by determining which lipids change in abundance during the ischemic process (Hankin et al, 2011; Koizumi et al, 2010; Wang et al, 2010) and the subsequent recovery (Whitehead et al, 2011). MALDI-MS imaging has also been used to explore the distribution of metabolites in a mouse ischemia model (middle-cerebral artery occlusion) (Hattori et al, 2010). TOF-SIMS has been used to image sodium, potassium, magnesium, and calcium ions in ischemic retinal tissues. Changes in the abundance of these ions were traced during ischemia progression, revealing that the abundance of calcium changed much more substantially over time than did that of the other studied ions (Kim et al, 2008). This technique was also used to quantify and image aluminum in human brain sections, demonstrating that it was much more abundant in the cerebral cortexes of chronic renal dialysis patients than in those of control patients (Candy et al, 1992).
Epilepsia
Changes in the distributions and abundance of energy-related metabolites were investigated in a murine model of kainate-induced limbic seizure (Sugiura et al, 2011). The results obtained indicate that CA3–cells consume energy selectively during seizures, and that there is an accelerated production of ATP within the brain to compensate for the resulting depletion. The abundance of ATP decreased massively during the induced seizures and that of ADP fell moderately, but there were no significant changes in the levels of AMP. A metabolomic analysis of energy synthesis pathway activity during the induced seizures revealed that they were associated with an acceleration of glycolysis and possibly TCA cycle activity, probably due to the depletion of ATP (Sugiura et al, 2011).
Psychiatric disorders
Surprisingly few studies have used MS imaging to study psychiatric disease states. A study that used MALDI-MS imaging to characterize lipids associated with schizophrenia demonstrated that phosphatidylcholine is abnormally distributed in the cortical layer of the frontal cortex in brain tissue samples from schizophrenic patients (Matsumoto et al, 2011). To find potential biomarkers for addiction, sections from nucleus accumbens that had developed behavioral sensitization to cocaine or corresponding saline-treated controls were analyzed with MALDI-MS imaging (Uys et al, 2010). Eight spectral features were found to be altered by cocaine treatment of which one was identified as secretoneurin (m/z 3653) having an increased expression.
FUTURE RESEARCH DIRECTIONS
Given the obvious importance of multimodal biomolecules and improved coverage of physical and chemical properties of molecules in the maintenance of good health and the progression of diseases, the ongoing development of novel tools that can be used to study these compounds will be essential in enabling us to understand disease pathogenesis and progression, and to develop effective treatments. The introduction and refinement of methods such as MS imaging makes it possible to conduct detailed studies on biological samples at the molecular level. The results discussed in this review clearly demonstrate that MS imaging is a powerful tool with the unique potential to enable comprehensive, label-free imaging of complex neural tissues. Moreover, the availability of both targeted and non-targeted methods of molecular imaging significantly expands the scope of imaging experiments.
MS imaging has a number of important advantages in neuroscience research. It is a technique for integrating biomolecular information that also provides chemical insights into the biomolecular associations between different groups of chemicals in neural tissues and shows how these associations can be affected by disease or the administration of a drug. However, MS imaging data sets are extremely complex as they consist of hundreds or thousands of molecule-specific images. Importantly, techniques have been developed that make it possible to overlay these images onto histological images obtained from the same tissue section, enabling the identification of correlations between changes in the abundance and localization of specific compounds in tissue samples with a high spatial resolution. MALDI-MS imaging can generate images with a spatial resolution of a few μm, whereas TOF-SIMS can achieve sub-μm resolutions. The power and capabilities of MS imaging technologies have increased substantially over the last decade, but further improvements are still needed, particularly with respect to sensitivity, quantitation, spatial resolution, the analysis of high molecular weight substances, wider coverage of multimodal molecules, compound identification, and throughput. Such improvements can be confidently expected because the technique is still young and is undergoing rapid development. To date, most of the publications describing its use have been reports of proof-of-principle and method development studies, and so there is considerable scope for its broader application in neuroscience research. It seems that MS imaging has the potential to find numerous uses in this field due to its ability as a tool for visualizing the distribution of important chemical species in biological samples and for providing quantitative data on their abundance.
FUNDING AND DISCLOSURE
We declare that, except for income received from our primary employer, no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
References
Ageta H, Asai S, Sugiura Y, Goto-Inoue N, Zaima N, Setou M (2009). Layer-specific sulfatide localization in rat hippocampus middle molecular layer is revealed by nanoparticle-assisted laser desorption/ionization imaging mass spectrometry. Med Mol Morphol 42: 16–23.
Angel PM, Spraggins JM, Baldwin HS, Caprioli R (2012). Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal Chem 84: 1557–1564.
Astigarraga E, Barreda-Gomez G, Lombardero L, Fresnedo O, Castano F, Giralt MT et al (2008). Profiling and imaging of lipids on brain and liver tissue by matrix-assisted laser desorption/ionization mass spectrometry using 2-mercaptobenzothiazole as a matrix. Anal Chem 80: 9105–9114.
Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A et al (2012). The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science 336: 1168–1171.
Bayes A, Grant SG (2009). Neuroproteomics: understanding the molecular organization and complexity of the brain. Nat Rev Neurosci 10: 635–646.
Becker JS, Dobrowolska J, Zoriy M, Matusch A (2008). Imaging of uranium on rat brain sections using laser ablation inductively coupled plasma mass spectrometry: a new tool for the study of critical substructures affined to heavy metals in tissues. Rapid Commun Mass Spectrom 22: 2768–2772.
Becker JS, Zoriy M, Becker JS, Dobrowolska J, Dehnhardt M, Matusch A (2007). Elemental imaging mass spectrometry of thin sections of tissues and analysis of brain proteins in gels by laser ablation inductively coupled plasma mass spectrometry. Phys Status Solidi C 4: 1775–1784.
Becker JS, Zoriy MV, Pickhardt C, Palomero-Gallagher N, Zilles K (2005). Imaging of copper, zinc, and other elements in thin section of human brain samples (Hippocampus) by laser ablation inductively coupled plasma mass spectrometry. Anal Chem 77: 3208–3216.
Benabdellah F, Seyer A, Quinton L, Touboul D, Brunelle A, Laprevote O (2010). Mass spectrometry imaging of rat brain sections: nanomolar sensitivity with MALDI versus nanometer resolution by TOF-SIMS. Anal Bioanal Chem 396: 151–162.
Benabdellah F, Touboul D, Brunelle A, Laprevote O (2009). In situ primary metabolites localization on a rat brain section by chemical mass spectrometry imaging. Anal Chem 81: 5557–5560.
Bezard E, Brotchie JM, Gross CE (2001). Pathophysiology of levodopa-induced dyskinesia: potential for new therapies. Nat Rev Neurosci 2: 577–588.
Bonnel D, Longuespee R, Franck J, Roudbaraki M, Gosset P, Day R et al (2011). Multivariate analyses for biomarkers hunting and validation through on-tissue bottom-up or in-source decay in MALDI-MSI: application to prostate cancer. Anal Bioanal Chem 401: 149–165.
Borner K, Nygren H, Hagenhoff B, Malmberg P, Tallarek E, Mansson JE (2006). Distribution of cholesterol and galactosylceramide in rat cerebellar white matter. Biochim Biophys Acta 1761: 335–344.
Burnum KE, Frappier SL, Caprioli RM (2008). Matrix-assisted laser desorption/ionization imaging mass spectrometry for the investigation of proteins and peptides. Annu Rev Anal Chem 1: 689–705.
Calligaris D, Longuespee R, Debois D, Asakawa D, Turtoi A, Castronovo V et al (2013). Selected protein monitoring in histological sections by targeted MALDI-FTICR in-source decay imaging. Anal Chem 85: 2117–2126.
Candy JM, Oakley AE, Mountfort SA, Taylor GA, Morris CM, Bishop HE et al (1992). The imaging and quantification of aluminum in the human brain using dynamic secondary ion mass-spectrometry (SIMS). Biol Cell 74: 109–118.
Caprioli RM, Farmer TB, Gile J (1997). Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 69: 4751–4760 The original MALDI-MS imaging publication demonstrating its usefulness to generate ion images of molecules in the surface layers of the tissue on biological tissue section.
Carter CL, McLeod CW, Bunch J (2011). Imaging of phospholipids in formalin fixed rat brain sections by matrix assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom 22: 1991–1998.
Castaing R, Slodzian G (1962). Microanalyse par émission ionique secondaire. J Microsc 1: 395–410.
Cerruti CD, Benabdellah F, Laprevote O, Touboul D, Brunelle A (2012). MALDI imaging and structural analysis of rat brain lipid negative ions with 9-aminoacridine matrix. Anal Chem 84: 2164–2171.
Cerruti CD, Touboul D, Guerineau V, Petit VW, Laprevote O, Brunelle A (2011). MALDI imaging mass spectrometry of lipids by adding lithium salts to the matrix solution. Anal Bioanal Chem 401: 75–87.
Chacon A, Zagol-Ikapitte I, Amarnath V, Reyzer ML, Oates JA, Caprioli RM et al (2011). On-tissue chemical derivatization of 3-methoxysalicylamine for MALDI-imaging mass spectrometry. J Mass Spectrom 46: 840–846.
Chan K, Lanthier P, Liu X, Sandhu JK, Stanimirovic D, Li JJ (2009). MALDI mass spectrometry imaging of gangliosides in mouse brain using ionic liquid matrix. Anal Chim Acta 639: 57–61.
Chaurand P, Schwartz SA, Caprioli RM (2002). Imaging mass spectrometry: a new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr Opin Chem Biol 6: 676–681.
Chen YF, Allegood J, Liu Y, Wang E, Cachon-Gonzalez B, Cox TM et al (2008). Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal Chem 80: 2780–2788.
Chochina SV, Avdulov NA, Igbavboa U, Cleary JP, O'Hare EO, Wood WG (2001). Amyloid beta-peptide1-40 increases neuronal membrane fluidity: role of cholesterol and brain region. J Lipid Res 42: 1292–1297.
Cohen LH, Gusev AI (2002). Small molecule analysis by MALDI mass spectrometry. Anal Bioanal Chem 373: 571–586.
Colsch B, Woods AS (2010). Localization and imaging of sialylated glycosphingolipids in brain tissue sections by MALDI mass spectrometry. Glycobiology 20: 661–667.
Cossec JC, Marquer C, Panchal M, Lazar AN, Duyckaerts C, Potier MC (2010). Cholesterol changes in Alzheimer’s disease: methods of analysis and impact on the formation of enlarged endosomes. Biochim Biophys Acta 1801: 839–845.
Debois D, Bertrand V, Quinton L, De Pauw-Gillet MC, De Pauw E (2010). MALDI-in source decay applied to mass spectrometry imaging: a new tool for protein identification. Anal Chem 82: 4036–4045.
Debois D, Brunelle A, Laprevote O (2007). Attempts for molecular depth profiling directly on a rat brain tissue section using fullerene and bismuth cluster ion beams. Int J Mass Spectrom 260: 115–120.
Delvolve AM, Woods AS (2011). Optimization of automated matrix deposition for biomolecular mapping using a spotter. J Mass Spectrom 46: 1046–1050.
Dill AL, Eberlin LS, Costa AB, Ifa DR, Cooks RG (2011). Data quality in tissue analysis using desorption electrospray ionization. Anal Bioanal Chem 401: 1949–1961.
Dobrowolska J, Dehnhardt M, Matusch A, Zoriy M, Palomero-Gallagher N, Koscielniak P et al (2008). Quantitative imaging of zinc, copper and lead in three distinct regions of the human brain by laser ablation inductively coupled plasma mass spectrometry. Talanta 74: 717–723.
Double KL, Gerlach M, Schunemann V, Trautwein AX, Zecca L, Gallorini M et al (2003). Iron-binding characteristics of neuromelanin of the human substantia nigra. Biochem Pharmacol 66: 489–494.
Eberlin LS, Dill AL, Golby AJ, Ligon KL, Wiseman JM, Cooks RG et al (2010a). Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew Chem Int Ed Engl 49: 5953–5956.
Eberlin LS, Ifa DR, Wu C, Cooks RG (2010b). Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew Chem Int Ed Engl 49: 873–876.
Ferguson CN, Benchaar SA, Miao Z, Loo JA, Chen H (2011). Direct ionization of large proteins and protein complexes by desorption electrospray ionization-mass spectrometry. Anal Chem 83: 6468–6473.
Franck J, Arafah K, Barnes A, Wisztorski M, Salzet M, Fournier I (2009). Improving tissue preparation for matrix-assisted laser desorption ionization mass spectrometry imaging. Part 1: Using microspotting. Anal Chem 81: 8193–8202.
Goodwin RJ, Dungworth JC, Cobb SR, Pitt AR (2008). Time-dependent evolution of tissue markers by MALDI-MS imaging. Proteomics 8: 3801–3808.
Goodwin RJ, Iverson SL, Andren PE (2012). The significance of ambient-temperature on pharmaceutical and endogenous compound abundance and distribution in tissues sections when analyzed by matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun Mass Spectrom 26: 494–498.
Goodwin RJ, Lang AM, Allingham H, Boren M, Pitt AR (2010). Stopping the clock on proteomic degradation by heat treatment at the point of tissue excision. Proteomics 10: 1751–1761.
Goodwin RJ, Mackay CL, Nilsson A, Harrison DJ, Farde L, Andren PE et al (2011). Qualitative and quantitative MALDI imaging of the positron emission tomography ligands raclopride (a D2 dopamine antagonist) and SCH 23390 (a D1 dopamine antagonist) in rat brain tissue sections using a solvent-free dry matrix application method. Anal Chem 83: 9694–9701.
Goto-Inoue N, Hayasaka T, Zaima N, Kashiwagi Y, Yamamoto M, Nakamoto M et al (2010). The detection of glycosphingolipids in brain tissue sections by imaging mass spectrometry using gold nanoparticles. J Am Soc Mass Spectrom 21: 1940–1943.
Groseclose MR, Andersson M, Hardesty WM, Caprioli RM (2007). Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom 42: 254–262.
Han XL, Holtzman DM, McKeel DW, Kelley J, Morris JC (2002). Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer’s disease: potential role in disease pathogenesis. J Neurochem 82: 809–818.
Hankin JA, Barkley RM, Murphy RC (2007). Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom 18: 1646–1652.
Hankin JA, Farias SE, Barkley R, Heidenreich K, Frey LC, Hamazaki K et al (2011). MALDI mass spectrometric imaging of lipids in rat brain injury models. J Am Soc Mass Spectrom 22: 1014–1021.
Hanrieder J, Ekegren T, Andersson M, Bergquist J (2013). MALDI imaging of post-mortem human spinal cord in amyotrophic lateral sclerosis. J Neurochem 124: 695–707.
Hanrieder J, Ljungdahl A, Falth M, Mammo SE, Bergquist J, Andersson M (2011). L-DOPA-induced dyskinesia is associated with regional increase of striatal dynorphin peptides as elucidated by imaging mass spectrometry. Mol Cell Proteomics 10: M111.009308.
Hardiman O, van den Berg LH, Kiernan MC (2011). Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol 7: 639–649.
Hardy J, Allsop D (1991). Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol Sci 12: 383–388.
Hardy J, Selkoe DJ (2002). The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297: 353–356.
Hare D, Reedy B, Grimm R, Wilkins S, Volitakis I, George JL et al (2009). Quantitative elemental bio-imaging of Mn, Fe, Cu and Zn in 6-hydroxydopamine induced Parkinsonism mouse models. Metallomics 1: 53–58 Elevated concentrations of elemental of Mn, Fe, Cu and Zn metals within the substantia nigra were imaged quantitated in Parkinson's disease mouse model brains using LA-ICP-MS.
Hattori K, Kajimura M, Hishiki T, Nakanishi T, Kubo A, Nagahata Y et al (2010). Paradoxical ATP elevation in ischemic penumbra revealed by quantitative imaging mass spectrometry. Antioxid Redox Signal 13: 1157–1167 Quantitative metabolomics revealed spatio-temporal changes in adenylates and NADH in a mouse middle-cerebral artery occlusion model.
Hillenkamp F, Unsold E, Kaufmann R, Nitsche R (1975). Laser microprobe mass analysis of organic materials. Nature 256: 119–120.
Hirsch-Reinshagen V, Burgess BL, Wellington CL (2009). Why lipids are important for Alzheimer disease? Mol Cell Biochem 326: 121–129.
Hsieh Y, Casale R, Fukuda E, Chen J, Knemeyer I, Wingate J et al (2006). Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Commun Mass Spectrom 20: 965–972.
Ifa DR, Wiseman JM, Song QY, Cooks RG (2007). Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI). Int J Mass Spectrom 259: 8–15 The original report on DESI-MS imaging demonstrating the feasibility of per-forming surface imaging studies of different chemicals and endogenous compounds.
Jehl B, Bauer R, Dorge A, Rick R (1981). The use of propane/isopentane mixtures for rapid freezing of biological specimens. J Microsc 123: 307–309.
Kallback P, Shariatgorji M, Nilsson A, Andren PE (2012). Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J Proteomics 75: 4941–4951 Protocols and software for the quantitation of drugs and endogenous compounds were demonstrated and the neuropeptide substance P was imaged, quantified and identified directly in different mouse brain structures using MALDI-MS.
Kim JH, Kim JH, Ahn BJ, Park JH, Shon HK, Yu YS et al (2008). Label-free calcium imaging in ischemic retinal tissue by TOF-SIMS. Biophys J 94: 4095–4102.
Koizumi S, Yamamoto S, Hayasaka T, Konishi Y, Yamaguchi-Okada M, Goto-Inoue N et al (2010). Imaging mass spectrometry revealed the production of lyso-phosphatidylcholine in the injured ischemic rat brain. Neuroscience 168: 219–225.
Komatsu M, Murayama Y, Hashimoto H (2008). Protein fragment imaging using ink jet printing digestion technique. Appl Surf Sci 255: 1162–1164.
Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ (2012). Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem 84: 141–148.
Lazar AN, Bich C, Panchal M, Desbenoit N, Petit VW, Touboul D et al (2013). Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients. Acta Neuropathol 125: 133–144 Original imaging approach to quantify cholesterol distribution in Alzheimer disease, showing that the cholesterol signal was significantly higher in the lower half of the cortex, layers III and IV, in AD samples compared to controls.
Lees AJ, Hardy J, Revesz T (2009). Parkinson’s disease. Lancet 373: 2055–2066.
Lemaire R, Wisztorski M, Desmons A, Tabet JC, Day R, Salzet M et al (2006). MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal Chem 78: 7145–7153.
Liebl H (1967). Ion microprobe mass analyzer. J Appl Phys 38: 5277–5283.
Ljungdahl A, Hanrieder J, Falth M, Bergquist J, Andersson M (2011). Imaging mass spectrometry reveals elevated nigral levels of dynorphin neuropeptides in L-DOPA-induced dyskinesia in rat model of Parkinson’s disease. PLoS ONE 6: e25653.
MacAleese L, Stauber J, Heeren RM (2009). Perspectives for imaging mass spectrometry in the proteomics landscape. Proteomics 9: 819–834 A concise review of MS imaging as a discovery oriented tool for proteomics as an alternative or complementary proteomics technique.
Manicke NE, Wiseman JM, Ifa DR, Cooks RG (2008). Desorption electrospray ionization (DESI) mass Spectrometry and tandem mass spectrometry (MS/MS) of phospholipids and sphingolipids: Ionization, adduct formation, and fragmentation. J Am Soc Mass Spectrom 19: 531–543.
Matsumoto J, Sugiura Y, Yuki D, Hayasaka T, Goto-Inoue N, Zaima N et al (2011). Abnormal phospholipids distribution in the prefrontal cortex from a patient with schizophrenia revealed by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal Bioanal Chem 400: 1933–1943.
Matusch A, Depboylu C, Palm C, Wu B, Hoglinger GU, Schafer MKH et al (2010). Cerebral bioimaging of Cu, Fe, Zn, and Mn in the MPTP mouse model of Parkinson’s disease using laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS). J Am Soc Mass Spectrom 21: 161–171.
McLean JA, Ridenour WB, Caprioli RM (2007). Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom 42: 1099–1105.
Meriaux C, Franck J, Wisztorski M, Salzet M, Fournier I (2010). Liquid ionic matrixes for MALDI mass spectrometry imaging of lipids. J Proteomics 73: 1204–1218.
Monnot AD, Behl M, Ho SN, Zheng W (2011). Regulation of brain copper homeostasis by the brain barrier systems: effects of Fe-overload and Fe-deficiency. Toxicol Appl Pharm 256: 249–257.
Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G et al (2010). Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PloS One 5: e11411 Original quantitative MALDI-MS imaging study in biological tissue sections where a drug was mapped and the concentration determined within microstructures of tissue sections.
Nilsson A, Skold K, Sjogren B, Svensson M, Pierson J, Zhang X et al (2007). Increased striatal mRNA and protein levels of the immunophilin FKBP-12 in experimental Parkinson’s disease and identification of FKBP-12-binding proteins. J Proteome Res 6: 3952–3961.
Norris JL, Caprioli RM (2013). Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev 113: 2309–2342 A comprehensive review of MALDI-MS imaging.
Nygren H, Borner K, Hagenhoff B, Malmberg P, Mansson JE (2005). Localization of cholesterol, phosphocholine and galactosylceramide in rat cerebellar cortex with imaging TOF-SIMS equipped with a bismuth cluster ion source. Biochim Biophys Acta 1737: 102–110.
Nygren H, Malmberg P (2010). High-resolution imaging and proteomics of peptide fragments by TOF-SIMS. Proteomics 10: 1694–1698.
Pan N, Liu PY, Cui WD, Tang B, Shi JM, Chen H (2013). Highly efficient ionization of phosphopeptides at low pH by desorption electrospray ionization mass spectrometry. Analyst 138: 1321–1324.
Passarelli MK, Winograd N (2011). Characterizing in situ Glycerophospholipids with SIMS and MALDI Methodologies. Surf Interface Anal 43: 269–271.
Pernber Z, Richter K, Mansson JE, Nygren H (2007). Sulfatide with different fatty acids has unique distributions in cerebellum as imaged by time-of-flight secondary ion mass spectrometry (TOF-SIMS). Biochim Biophys Acta 1771: 202–209.
Perrin RJ, Fagan AM, Holtzman DM (2009). Multimodal techniques for diagnosis and prognosis of Alzheimer's disease. Nature 461: 916–922.
Pierson J, Norris JL, Aerni HR, Svenningsson P, Caprioli RM, Andren PE (2004). Molecular profiling of experimental Parkinson's disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J Proteome Res 3: 289–295 Histology-directed protein profiling was used for the first time to generate peptide and protein profiles and relative levels directly from brain tissue sections in a Parkinson's disease animal model.
Pierson J, Svenningsson P, Caprioli RM, Andren PE (2005). Increased levels of ubiquitin in the 6-OHDA-lesioned striatum of rats. J Proteome Res 4: 223–226 Histology-directed protein profiling of unconjugated ubiquitin levels in tissue sections.
Pirman DA, Reich RF, Kiss A, Heeren RMA, Yost RA (2013). Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal Chem 85: 1081–1089.
Pol J, Vidova V, Kruppa G, Kobliha V, Novak P, Lemr K et al (2009). Automated ambient desorption-ionization platform for surface imaging integrated with a commercial fourier transform ion cyclotron resonance mass spectrometer. Anal Chem 81: 8479–8487.
Przybylski C, Gonnet F, Hersant Y, Bonnaffe D, Lortat-Jacob H, Daniel R (2010). Desorption electrospray ionization mass spectrometry of glycosaminoglycans and their protein noncovalent complex. Anal Chem 82: 9225–9233.
Puglielli L, Tanzi RE, Kovacs DM (2003). Alzheimer’s disease: the cholesterol connection. Nat Neurosci 6: 345–351.
Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM (2008). Solvent-free matrix dry-coating for MALDI Imaging of phospholipids. J Am Soc Mass Spectrom 19: 882–886.
Qin ZY, Caruso JA, Lai B, Matusch A, Becker JS (2011). Trace metal imaging with high spatial resolution: applications in biomedicine. Metallomics 3: 28–37.
Rasia RM, Bertoncini CW, Marsh D, Hoyer W, Cherny D, Zweckstetter M et al (2005). Structural characterization of copper(II) binding to alpha-synuclein: insights into the bioinorganic chemistry of Parkinson’s disease. Proc Natl Acad Sci USA 102: 4294–4299.
Rohner TC, Staab D, Stoeckli M (2005). MALDI mass spectrometric imaging of biological tissue sections. Mech Ageing Dev 126: 177–185.
Schober Y, Schramm T, Spengler B, Rompp A (2011). Protein identification by accurate mass matrix-assisted laser desorption/ionization imaging of tryptic peptides. Rapid Commun Mass Spectrom 25: 2475–2483.
Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM (2008). Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am Soc Mass Spectrom 19: 1069–1077.
Sensi SL, Paoletti P, Koh JY, Aizenman E, Bush AI, Hershfinkel M (2011). The Neurophysiology and pathology of Brain Zinc. J Neurosci 31: 16076–16085.
Shanta SR, Zhou LH, Park YS, Kim YH, Kim Y, Kim KP (2011). Binary matrix for MALDI imaging mass spectrometry of phospholipids in both ion modes. Anal Chem 83: 1252–1259.
Shariatgorji M, Amini N, Ilag LL (2009). Silicon nitride nanoparticles for surface-assisted laser desorption/ionization of small molecules. J Nanopart Res 11: 1509–1512.
Shariatgorji M, Kallback P, Gustavsson L, Schintu N, Svenningsson P, Goodwin RJA et al (2012a). Controlled-pH tissue cleanup protocol for signal enhancement of small molecule drugs analyzed by MALDI-MS imaging. Anal Chem 84: 4603–4607.
Shariatgorji M, Nilsson A, Goodwin RJA, Svenningsson P, Schintu N, Banka Z et al (2012b). Deuterated matrix-assisted laser desorption ionization matrix uncovers masked mass spectrometry imaging signals of small molecules. Anal Chem 84: 7152–7157 A deuterated MALDI-MS matrix was developed to facilitate imaging of small molecule pharmaceuticals and endogenous compounds including the neurotransmitter acetylcholine directly from biological tissue sections.
Shiea J, Yuan CH, Huang MZ, Cheng SC, Ma YL, Tseng WL et al (2008). Detection of native protein ions in aqueous solution under ambient conditions by electrospray laser desorption/ionization mass spectrometry. Anal Chem 80: 4845–4852.
Shimma S, Sugiura Y, Hayasaka T, Zaima N, Matsumoto M, Setou M (2008). Mass imaging and identification of biomolecules with MALDI-QIT-TOF-based system. Anal Chem 80: 878–885.
Shrivas K, Hayasaka T, Goto-Inoue N, Sugiura Y, Zaima N, Setou M (2010). Ionic matrix for enhanced MALDI imaging mass spectrometry for identification of phospholipids in mouse liver and cerebellum tissue sections. Anal Chem 82: 8800–8806.
Shrivas K, Hayasaka T, Sugiura Y, Setou M (2011). Method for simultaneous imaging of endogenous low molecular weight metabolites in mouse brain using TiO2 nanoparticles in nanoparticle-assisted laser desorption/ionization-imaging mass spectrometry. Anal Chem 83: 7283–7289.
Sjovall P, Johansson B, Lausmaa J (2006). Localization of lipids in freeze-dried mouse brain sections by imaging TOF-SIMS. Appl Surf Sci 252: 6966–6974.
Sjovall P, Lausmaa J, Johansson B (2004). Mass spectrometric imaging of lipids in brain tissue. Anal Chem 76: 4271–4278.
Sjovall P, Lausmaa J, Nygren H, Carlsson L, Malmberg P (2003). Imaging of membrane lipids in single cells by imprint-imaging time-of-flight secondary ion mass spectrometry. Anal Chem 75: 3429–3434.
Skold K, Svensson M, Nilsson A, Zhang XQ, Nydahl K, Caprioli RM et al (2006). Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J Proteome Res 5: 262–269.
Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE (2007). The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: Stathmin 2-20 and peptides as sample quality indicators. Proteomics 7: 4445–4456.
Sloot WN, Gramsbergen JB (1994). Axonal transport of manganese and its relevance to selective n eurotoxicity in the rat basal ganglia. Brain Res 657: 124–132.
Sole-Domenech S, Sjovall P, Vukojevic V, Fernando R, Codita A, Salve S et al (2013). Localization of cholesterol, amyloid and glia in Alzheimer's disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (ToF-SIMS) and immunofluorescence imaging. Acta Neuropathol 125: 145–157 The spatial distributions of lipids, amyloid-beta deposits, markers of neurons and glial cells were imaged at sub-μm lateral resolution in brain structures of a mouse model of Alzheimer's disease and revealed cholesterol accumulations in hippocampal regions undergoing amyloid-beta deposition.
Stauber J, MacAleese L, Franck J, Claude E, Snel M, Kaletas BK et al (2010). On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J Am Soc Mass Spectrom 21: 338–347.
Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM (2001). Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med 7: 493–496.
Stoeckli M, Knochenmuss R, McCombie G, Mueller D, Rohner T, Staab D et al (2006). MALDI MS imaging of amyloid. Methods Enzymol 412: 94–106.
Stoeckli M, Staab D, Staufenbiel M, Wiederhold KH, Signor L (2002). Molecular imaging of amyloid beta peptides in mouse brain sections using mass spectrometry. Anal Biochem 311: 33–39 MALDI-MS imaging was applied to the study of amyloid beta peptide distribution in brain sections from an Alzheimer's disease mouse model.
Sugiura Y, Konishi Y, Zaima N, Kajihara S, Nakanishi H, Taguchi R et al (2009a). Visualization of the cell-selective distribution of PUFA-containing phosphatidylcholines in mouse brain by imaging mass spectrometry. J Lipid Res 50: 1776–1788.
Sugiura Y, Setou M (2009b). Selective imaging of positively charged polar and nonpolar lipids by optimizing matrix solution composition. Rapid Commun Mass Spectrom 23: 3269–3278.
Sugiura Y, Taguchi R, Setou M (2011). Visualization of spatiotemporal energy dynamics of hippocampal neurons by mass spectrometry during a kainate-induced seizure. PLoS ONE 6: e17952.
Sugiura Y, Zaima N, Setou M, Ito S, Yao I (2012). Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal Bioanal Chem 403: 1851–1861.
Svensson M, Boren M, Skold K, Falth M, Sjogren B, Andersson M et al (2009). Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res 8: 974–981.
Svensson M, Skold K, Nilsson A, Falth M, Nydahl K, Svenningsson P et al (2007). Neuropeptidomics: MS applied to the discovery of novel peptides from the brain. Anal Chem 79: 15–16 18-21.
Svensson M, Skold K, Svenningsson P, Andren PE (2003). Peptidomics-based discovery of novel neuropeptides. J Proteome Res 2: 213–219.
Takats Z, Wiseman JM, Gologan B, Cooks RG (2004). Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306: 471–473 The first report of DESI-MS demonstrating that electrosprayed charged droplets and ions of solvent could be directed onto a tissue surface to produce gaseous ions of material originally present on the surface.
Thomas A, Charbonneau JL, Fournaise E, Chaurand P (2012). Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal Chem 84: 2048–2054.
Touboul D, Halgand F, Brunelle A, Kersting R, Tallarek E, Hagenhoff B et al (2004). Tissue molecular ion imaging by gold cluster ion bombardment. Anal Chem 76: 1550–1559.
Trim PJ, Atkinson SJ, Princivalle AP, Marshall PS, West A, Clench MR (2008). Matrix-assisted laser desorption/ionisation mass spectrometry imaging of lipids in rat brain tissue with integrated unsupervised and supervised multivariant statistical analysis. Rapid Commun Mass Spectrom 22: 1503–1509.
Trim PJ, Djidja MC, Atkinson SJ, Oakes K, Cole LM, Anderson DM et al (2010). Introduction of a 20 kHz Nd:YVO4 laser into a hybrid quadrupole time-of-flight mass spectrometer for MALDI-MS imaging. Anal Bioanal Chem 397: 3409–3419.
Trimpin S, Herath TN, Inutan ED, Wager-Miller J, Kowalski P, Claude E et al (2010). Automated solvent-free matrix deposition for tissue imaging by mass spectrometry. Anal Chem 82: 359–367.
Uys JD, Grey AC, Wiggins A, Schwacke JH, Schey KL, Kalivas PW (2010). Matrix-assisted laser desorption/ionization tissue profiling of secretoneurin in the nucleus accumbens shell from cocaine-sensitized rats. J Mass Spectrom 45: 97–103.
Wang HYJ, Liu CB, Wu HW, Kuo S (2010). Direct profiling of phospholipids and lysophospholipids in rat brain sections after ischemic stroke. Rapid Commun Mass Spectrom 24: 2057–2064.
Wang HYJ, SNJJ Post, Woods AS (2008). A minimalist approach to MALDI imaging of glycerophospholipids and sphingolipids in rat brain sections. Int J Mass Spectrom 278: 143–149.
Wei J, Buriak JM, Siuzdak G (1999). Desorption-ionization mass spectrometry on porous silicon. Nature 399: 243–246.
Wenk GL (2003). Neuropathologic changes in Alzheimer’s disease. J Clin Psychiatry 64 (Suppl 9): 7–10.
Whitehead SN, Chan KH, Gangaraju S, Slinn J, Li J, Hou ST (2011). Imaging mass spectrometry detection of gangliosides species in the mouse brain following transient focal cerebral ischemia and long-term recovery. PLoS ONE 6: e20808.
Wiseman JM, Ifa DR, Song QY, Cooks RG (2006). Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew Chem Int Ed Engl 45: 7188–7192.
Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT et al (2008). Desorption electrospray ionization mass spectrometry: imaging drugs and metabolites in tissues. Proc Natl Acad Sci USA 105: 18120–18125.
Wu CP, Ifa DR, Manicke NE, Cooks RG (2009). Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal Chem 81: 7618–7624.
Wu CP, Ifa DR, Manicke NE, Cooks RG (2010). Molecular imaging of adrenal gland by desorption electrospray ionization mass spectrometry. Analyst 135: 28–32 Subtle differences in the spatial distributions of the catecholamines epinephrine and norepinephrine were distinguished in tissue sections from porcine adrenal glands.
Yang HJ, Ishizaki I, Sanada N, Zaima N, Sugiura Y, Yao I et al (2010a). Detection of characteristic distributions of phospholipid head groups and fatty acids on neurite surface by time-of-flight secondary ion mass spectrometry. Med Mol Morphol 43: 158–164.
Yang HJ, Sugiura Y, Ishizaki I, Sanada N, Ikegami K, Zaima N et al (2010b). Imaging of lipids in cultured mammalian neurons by matrix assisted laser/desorption ionization and secondary ion mass spectrometry. Surf Interface Anal 42: 1606–1611.
Yang J, Caprioli RM (2013). Matrix precoated targets for direct lipid analysis and imaging of tissue. Anal Chem 85: 2907–2912.
Yuki D, Sugiura Y, Zaima N, Akatsu H, Hashizume Y, Yamamoto T et al (2011). Hydroxylated and non-hydroxylated sulfatide are distinctly distributed in the human cerebral cortex. Neuroscience 193: 44–53.
Zecca L, Youdim MBH, Riederer P, Connor JR, Crichton RR (2004). Iron, brain ageing and neurodegenerative disorders. Nat Rev Neurosci 5: 863–873.
Zoriy MV, Becker JS (2007). Imaging of elements in thin cross sections of human brain samples by LA-ICP-MS: a study on reproducibility. Int J Mass Spectrom 264: 175–180.
Zoriy MV, Dehnhardt M, Matusch A, Becker JS (2008). Comparative imaging of P, S, Fe, Cu, Zn and C in thin sections of rat brain tumor as well as control tissues by laser ablation inductively coupled plasma mass spectrometry. Spectrochim Acta B 63: 375–382.
Zoriy MV, Dehnhardt M, Reifenberger G, Zilles K, Becker JS (2006). Imaging of Cu, Zn, Pb and U in human brain tumor resections by laser ablation inductively coupled plasma mass spectrometry. Int J Mass Spectrom 257: 27–33.
Acknowledgements
This work was supported by the Swedish Research Council, Grant 2011–2013, 2010–2016, 2011–2014 and 2010–2014.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shariatgorji, M., Svenningsson, P. & Andrén, P. Mass Spectrometry Imaging, an Emerging Technology in Neuropsychopharmacology. Neuropsychopharmacol 39, 34–49 (2014). https://doi.org/10.1038/npp.2013.215
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2013.215
Keywords
This article is cited by
-
MALDI imaging mass spectrometry: an emerging tool in neurology
Metabolic Brain Disease (2022)
-
Spatial visualization of comprehensive brain neurotransmitter systems and neuroactive substances by selective in situ chemical derivatization mass spectrometry imaging
Nature Protocols (2021)
-
Neuropharmacokinetic visualization of regional and subregional unbound antipsychotic drug transport across the blood–brain barrier
Molecular Psychiatry (2021)
-
Molecular imaging identifies age-related attenuation of acetylcholine in retrosplenial cortex in response to acetylcholinesterase inhibition
Neuropsychopharmacology (2019)
-
Spatial and molecular changes of mouse brain metabolism in response to immunomodulatory treatment with teriflunomide as visualized by MALDI-MSI
Analytical and Bioanalytical Chemistry (2019)