Abstract
Cocaine dependence (CD) and major depressive episode (MDE) frequently co-occur with poorer treatment outcome and higher relapse risk. Shared genetic risk was affirmed; to date, there have been no reports of genomewide linkage scans (GWLSs) surveying the susceptibility regions for comorbid CD and MDE (CD–MDE). We aimed to identify chromosomal regions and candidate genes susceptible to CD, MDE, and CD–MDE in African Americans (AAs) and European Americans (EAs). A total of 1896 individuals were recruited from 384 AA and 355 EA families, each with at least a sibling-pair with CD and/or opioid dependence. Array-based genotyping of about 6000 single-nucleotide polymorphisms was completed for all individuals. Parametric and non-parametric genomewide linkage analyses were performed. We found a genomewide-significant linkage peak on chromosome 7 at 183.4 cM for non-parametric analysis of CD–MDE in AAs (lod=3.8, genomewide empirical p=0.016; point-wise p=0.00001). A nearly genomewide significant linkage was identified for CD–MDE in EAs on chromosome 5 at 14.3 cM (logarithm of odds (lod)=2.95, genomewide empirical p=0.055; point-wise p=0.00012). Parametric analysis corroborated the findings in these two regions and improved the support for the peak on chromosome 5 so that it reached genomewide significance (heterogeneity lod=3.28, genomewide empirical p=0.046; point-wise p=0.00053). This is the first GWLS for CD–MDE. The genomewide significant linkage regions on chromosomes 5 and 7 harbor four particularly promising candidate genes: SRD5A1, UBE3C, PTPRN2, and VIPR2. Replication of the linkage findings in other populations is warranted, as is a focused analysis of the genes located in the linkage regions implicated here.
Similar content being viewed by others
INTRODUCTION
Major depression and cocaine dependence (CD) frequently co-occur. Major depression ranks as one of the top three mental disorders in terms of burden of disease (Evans and Charney, 2003), and CD ranks as the third most prevalent illicit drug dependence diagnosis (SAMSHA, 2009). There is a high rate of comorbidity between CD and depression observed clinically. Although there are no reported statistics for prevalence of comorbid CD and depression, it is estimated that approximately 30% of people with major depressive disorder have a lifetime drug use disorder, and the lifetime prevalence of affective disorders is about 35% among cocaine abusers (Regier et al, 1990). Similar comorbidity rates were observed in the Sequenced Treatment Alternatives to Relieve Depression study (Davis et al, 2006) and in the 2008 National Survey on Drug Use and Health (SAMSHA, 2009). This comorbidity is associated with a higher risk of suicide, greater social and functional impairment, and greater risk of other co-occurring psychiatric disorders (Davis et al, 2006). ‘The family data used for this study show that CD–major depressive episode (MDE) subjects’ relatives have specifically increased risk of CD–MDE. The probability of at least two siblings with CD–MDE among all the families with CD–MDE is 0.28 for African American (AA) and 0.37 for European American (EA), both of which are higher than the prevalence in the general population. In the United States, the lifetime prevalence of MDD was estimated by the National Health and Nutrition Examination Survey III (Riolo et al, 2005) to be 0.104 and 0.075 for EA and AA, respectively. Although prevalence estimates are not available for CD–MDE, the trait for which we aimed to map susceptibility genomic regions in this study, it has a lower population prevalence than CD alone by virtue of being a subset of those affected with the disorder.
Evidence from adoption, twin, and family studies supports the important role of genetic factors in vulnerability to comorbid depression and substance dependence (Ingraham and Wender, 1992; Prescott et al, 2000; Maher et al, 2002). Estimates from twin studies of the heritability of CD are as high as 72% for CD (Goldman et al, 2005), and as high as 58% for depression (Uhl and Grow, 2004).
The first genomewide linkage scan (GWLS) of comorbid alcoholism and depression (Nurnberger et al, 2001) was published by the Collaborative Study of the Genetics of Alcoholism research group. Although they found significant logarithm of odds (lod) scores on four chromosomes, the significant signals each appeared in only one of the two data sets. They reported that chromosome 1, near 120 centimorgans (cM), might harbor genes for the alcoholism or depression phenotypes with lod scores of 5.12 and 1.52 in the two data sets, respectively (4.66 in the combined data set) and concluded that a locus on chromosome 1 influences vulnerability to alcoholism and affective disorder.
Genomewide association studies (GWAS) have been widely employed in the last few years to identify risk genes for complex traits. Many risk loci have been identified, but loci found through the GWAS generally account for only a small amount of the genetic risk for the trait, typically with an odds ratio of 1.2 or lower for each locus (Manolio et al, 2009). More recently, it has been recognized that multiple rare variants at risk loci may account for some of the ‘missing’ heritability (Manolio et al, 2009; Eichler et al, 2010). Such disease-associated rare variants are unlikely to be detected through GWAS because of the low correlation between common single-nucleotide polymorphisms (SNPs) and rare variants, but an adequately powered genetic linkage analysis could detect their effects (McMillan and Robertson, 1974; Ng et al, 2009), because segregation of multiple causal rare variants at the same locus can sum to a detectable linkage signal over a sample. Therefore, linkage studies are still a useful method for gene discovery. To our knowledge, there have been no previous GWLS reports for the comorbidity of CD and depression, and specific genes associated with comorbid CD and depression have yet to be identified. To address this issue, we conducted an autosomal GWLS for comorbid CD and MDE to identify genomic regions likely to contain risk loci for the co-occurrence of the two disorders.
MATERIALS AND METHODS
Recruitment
Subjects were initially ascertained for genetic studies of CD and opioid dependence (OD) (Gelernter et al, 2006) using the affected sibling pair (ASP) linkage approach. There were four recruitment sites: Yale University School of Medicine (APT Foundation; New Haven, Connecticut), University of Connecticut Health Center (UConn; Farmington, Connecticut), Medical University of South Carolina (MUSC; Charleston, South Carolina), and McLean Hospital (Harvard Medical School; Belmont, Massachusetts). The recruitment and assessment were nearly identical between the two studies (other than for primary trait of interest), and the recruitment periods overlapped. Families were screened and recruited based on the belief that at least two siblings would meet diagnostic criteria for CD or OD. CD families were recruited from all sites, while OD families were recruited only at the UConn and Yale sites. The families used for this linkage study included all those with at least one CD sib-pair, which also included those comorbid with OD (but no OD families that did not also include at least one CD sib-pair). Probands with an axis I clinical diagnosis of a major psychotic disorder such as schizophrenia or schizoaffective disorder were excluded from participation. Regardless of affection status, other siblings and parents were recruited whenever available, to increase linkage information. After a complete description of the study was provided to the subjects, written informed consent was obtained from all subjects. The institutional review board at each recruitment site approved all study materials and the National Institute on Drug Abuse issued a certificate of confidentiality for the work.
Diagnosis and Study Subjects
Subjects were interviewed with the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al, 2005; Gelernter et al, 2005). In addition to a section on the diagnosis of CD, a section of the SSADDA is devoted to the assessment of depressive disorders. The diagnoses of CD and MDE were based on the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV). On the basis of DSM-IV, we drew a distinction between a MDE that is substance induced and one that is independent. However, we combined them to increase sample size, denoted as ‘MDE.’ An individual was considered affected for the comorbid CD and MDE (CD–MDE) only if diagnosed with both CD and MDE (not necessary at the same time). Table 1 presents the demographic characteristics of the sample, which was approximately evenly divided by sex and ranged in age from 18 to 61 (mean±SD=39.5±8.8). In all, 65% of the families had one or more affected relatives with a lifetime history of MDE, and 92.5% of the families that were positive for MDE had one or more family members who were comorbid for CD and MDE. Table 2 shows the number of informative families used in the GWLS for each trait.
Genotyping and Quality Control
DNA was obtained from immortalized cell lines in most cases, but for a small number of subjects DNA was obtained directly from blood or saliva. A total of 1630 individuals were genotyped at the Center for Inherited Disease Research for the 6008 SNPs Illumina Human Linkage IVb Marker Panel. An additional 266 individuals were genotyped at Yale (Keck Center) with the 6090 SNP Illumina Infinium-12 Human Linkage Marker Panel. We limited our analyses to the autosomal SNPs, and used 5636 and 5735 SNPs from the first and second panels, respectively. Among the SNPs in the two panels, 4518 SNPs were in common across the two platforms, and were used for the following quality control and analyses. PLINK software (Purcell et al, 2007) was used to calculate allele frequencies and examine Hardy–Weinberg equilibrium (HWE) in each population using a set of unrelated subjects (355 EA and 384 AA subjects were randomly selected, one per family). SNPs with a genotyping rate ⩽0.95, a minor allele frequency (MAF) ⩽0.1, or not in HWE (p⩽0.01) were excluded. We used PedCheck (O’Connell and Weeks, 1998) and Merlin (Abecasis et al, 2002) to identify Mendelian inconsistencies. We also used Merlin to identify potentially incorrect genotypes based on estimation of the probability of double-crossover events using the Merlin ‘––error’ option. We used the Pedigree RElationship Statistical Test (PREST) (McPeek and Sun, 2000) to verify family relationships.
There were 21(15), 318 (68), and 46 (40) SNPs that failed the criteria of genotyping rate, MAF and HWE in AAs (EAs), respectively. We limited our analyses to the 4133 (4395) remaining autosomal markers in AAs (EAs). A total of 168 (46) Mendelian inconsistencies were identified by PedCheck and possible genotyping error rates of 0.048% (0.079%) were suggested by Merlin in AAs (EAs). These problematic genotypes were set to be missing in linkage analysis. Pedigree errors were detected in two AA families and five EA families by PREST. Of these, the relationships in one AA family and five EA families were corrected based on the shared identical by decent (IBD) patterns and the re-assigned family relationships were verified by PREST. One AA family relationship could not be resolved and the family was excluded from further analysis. Population was assigned by individual genetic profile (refer to Supplementary File 1 for details).
Linkage Analyses
Initially, we used Merlin (Abecasis et al, 2002) and a non-parametric (ie, model free), penetrance-independent, affected-only, allele-sharing model to test for linkage. Allele frequencies were estimated by counting all genotyped individuals. The Kong and Cox linear allele-sharing model (Kong and Cox, 1997) was used to estimate the lod score.
We used Monte Carlo simulations under the null hypothesis of random linkage between phenotype and genotype to assess the empirical thresholds for genomewide suggestive and significant linkages. We used the gene-dropping algorithm implemented in Merlin to simulate 1000 data sets conditional on the observed family structure, marker spacing, allele frequencies, and missing data pattern. To reduce the computational burden attributable to modeling maker-marker LD, we used a screened marker set with low LD (3675 SNPs in AAs and 3760 SNPs in EAs), that is, r2<0.1 for each pair of markers. Each simulated data set was then analyzed using the same procedure as for the observed data. The highest lod score for each chromosome was recorded for each simulated data set. The genomewide suggestive linkage threshold is defined as the maximum lod score expected once by chance per genome scan (Lander and Kruglyak, 1995) and set as the 1000 highest lod score out of 22 000 lod scores for each of the 22 autosomal chromosomes from the 1000 simulations. The genomewide significant threshold was set as the 95th percentile of the distribution of 22 000 lod scores. These empirical thresholds are shown in Table 2. The autosomal genomewide empirical significance of an observed lod score was derived from the same simulation. It was estimated by counting how often the entire genome had a maximum lod score greater than or equal to the observed score across the 1000 simulated replicates.
Parametric linkage analysis may increase statistical power by including information contributed by families with only one affected and unaffected individuals. We conducted parametric linkage analysis using the program Merlin (Abecasis et al, 2002) to verify the significant linkage signals from non-parametric analysis and identify other linkage regions. To avoid missing evidence for linkage because of genetic model misspecification, the parameters used in the analyses covered a wide range of both dominant and recessive models: nine levels of penetrance from 0.1 to 0.9 with increment of 0.1 for the disease gene carriers, two levels of penetrance of 0.005 and 0.01 for the non-risk genotype, and fixed disease allele frequencies of 0.05 and 0.3 for the dominant and recessive models, respectively. Hence, 36 models were examined and correction for multiple testing was done using empirical simulations on a genomewide scale. We calculated heterogeneity lod (hlod) scores to allow for linkage heterogeneity using Merlin, which internally maximizes parameters to estimate the degree of heterogeneity. The maximum hlod score over the 36 examined genetic models is presented for each locus. The empirical thresholds of Lander–Kruglyak for suggestive and significant linkages (Lander and Kruglyak, 1995), and the empirical genomewide significance for the maximized hlod scores, were assessed by 1000 simulations in the same way as the non-parametric linkage analysis described above.
RESULTS
Comorbidity in the Sample
The presence of either CD or MDE increased the risk of the other disorder, as is commonly observed. More than one-third (37.4%) of 1896 participants or 39.1% of 739 families had co-occurring lifetime diagnoses of both CD and MDE. The presence of CD more than doubled the likelihood of a diagnosis of MDE (44.8% in those with CD vs 24.3% in those without CD). The presence of MDE also increased the risk of CD; of those affected with MDE, 90.3% had CD, compared with 78.5% of those without MDE. The high rates of CD, with or without MDE, reflect the fact that the majority of the sample was ascertained as CD sibling pairs.
Population Assignment
The marker-based population assignment resulted in the majority of self-reported AA and Hispanic black subjects clustering with the AA group, and the majority of self-reported white and Hispanic white subjects clustering with the EA group. Of the self-reported AAs, 5.7% were re-classified to EA. Of the self-reported EAs, 0.76% were re-classified to AA. For mixed-race families, 10.8% were assigned to AA and 15.3% to EA.
Non-Parametric Linkage Analysis
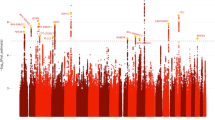
Figure 1 shows non-parametric linkage lod scores from the analysis of AAs. Table 2 lists details of the chromosomal regions that exceeded the empirical threshold for genomewide suggestive linkage for the three phenotypes in AAs. The strongest evidence for linkage reached genomewide significance with the highest lod score of 3.8 on chromosome 7 at 183.4 cM in the analysis of the comorbid CD–MDE phenotype (genomewide empirical p=0.016; point-wise p=0.00001). In the same region, we also observed a genomewide suggestive linkage in the analysis of MDE (lod=3.01, genomewide empirical p=0.065; point-wise p=0.0001), whereas no evidence for linkage was detected for CD. Five other linkage peaks exceeded the empirical genomewide suggestive linkage cutoffs: three peaks on chromosomes 3 (145.5 cM), 9 (99.8 cM), and 10 (144.4 cM) for CD, two peaks on chromosome 2 at 59.9 and 71.9 cM for MDE and CD–MDE, respectively.
Lod scores on 22 autosomal chromosomes resulting from non-parametric linkage analyses including African-American families with cocaine dependence (CD), major depressive episode (MDE), and comorbid CD with MDE (CD–MDE). The dashed lines denote the thresholds of lod scores for the three phenotypes for genomewide-‘suggestive’ linkage. (Some of these thresholds appear as one dashed line because they are very close).
The linkage peaks identified in the EA sample were different from those in the AA sample (Figure 2 and Table 2). A nearly genomewide significant linkage peak with lod=2.95 was observed on chromosome 5 at 14.3 cM for the comorbid CD–MDE phenotype (genomewide empirical p=0.055; point-wise p=0.00012), while at the same location there was only modest evidence for linkage for MDE (lod=1.52, point-wise p=0.004) and no evidence of linkage for CD. Four other chromosomal regions showed suggestive linkage signals: chromosomes 18 (110.5 cM), 13 (102.7 cM), and 16 (76.3 cM) for CD, and 10 (153.5 cM) for MDE, and 16 (76.3 cM) CD–MDE.
Lod scores on 22 autosomal chromosomes resulting from non-parametric linkage analyses including European-American families with cocaine dependence (CD), major depressive episode (MDE), and comorbid CD and MDE (CD–MDE). The dashed lines denote the thresholds of lod scores for the three phenotypes for genomewide suggestive linkage. (Some of these thresholds appear as one dashed line because they are very close).
Parametric Linkage Analysis
We performed parametric analysis to examine the robustness of the non-parametric results and potentially identify additional linkage peaks. In the AA sample, the linkage peaks identified by non-parametric analysis were confirmed (Figure 3). Linkage peaks for CD on chromosome 10 at 144.4 cM (peak hlod=4.05) and for MDE (peak hlod=3.60) and CD–MDE (peak hlod=4.58) on chromosome 7 at 183.4 cM attained genomewide significance (Supplementary Table 1 and Supplementary Figure S1). Additionally, a new suggestive linkage peak for CD was identified on chromosome 1 at 61.5 cM (peak hlod=2.04). In the EA sample, parametric analysis confirmed the suggestive linkage peaks for CD–MDE (Figure 3 and Supplementary Figure S2), and the peak on chromosome 5 at 15.3 cM (peak hlod=3.28) attained genomewide significance (Supplementary Table 1). Four new regions on chromosomes 21 (34.6 cM) for CD, 3 (0.98 cM) for MDE, 15 (92.9 cM) for MDE, and 9 (82.2 cM) for CD–MDE reached suggestive linkage thresholds. However, the two suggestive linkage regions on chromosomes 18 (110.5 cM) for CD) and 13 (102.7 cM) for MDE identified by the non-parametric approach did not reach genomewide suggestive linkage criteria in the parametric linkage analysis.
Hlod scores on chromosome 7 for African Americans and chromosome 5 for European Americans resulting from the parametric linkage analysis of all possible sibling pairs with cocaine dependence (CD), major depressive episode (MDE), and comorbid CD and MDE (CD–MDE). The dash lines denoted the thresholds of hlod scores for the three phenotypes for genomewide suggestive linkage. (Some of these thresholds appear as one dashed line because they are very close).
DISCUSSION
In this first GWLS for comorbid CD and MDE, we found genomewide significant evidence for loci on chromosome 5 in the EA families and chromosome 7 in the AA families. We also obtained genomewide significant evidence in the AA sample for linkage of CD to a novel locus on chromosome 10. This linkage peak is located 10–15 cM proximal to a suggestive linkage peak for CD–MDE observed in the EA families. Moreover, the chromosome 10 locus for CD in the AA families is 17 and 27 cM distal from linkage peaks we found for alcohol dependence (AD) previously in the EA and AA families, respectively (Gelernter et al, 2009; Panhuysen et al, 2010).
The linkage region on chromosome 5 for CD–MDE spans from 11.2 to 16.7 cM as defined by the 1-lod support interval, is predicted to contain 18 genes, and harbors a particularly promising candidate gene, SRD5A1. This gene encodes 5-alpha-reductase type I, which catalyzes the synthesis of the neurosteroid allopregnanolone, a potent positive modulator of gamma amino butyric acid (GABA) action at GABA-A receptors in the brain (Agís-Balboa et al, 2006). The de novo synthesis of neurosteroids in the brain during stress and after alcohol consumption has been reported (Reddy, 2003; Kumar et al, 2004), and disruption of steroid-regulated GABAergic inhibition has been implicated in anxiety, major depression, AD, and schizophrenia (Eser et al, 2006; MacKenzie et al, 2007; Tanchuck et al, 2009).
The linkage region on chromosome 7 is about 2 cM from the telomere of the long arm and harbors approximately 20 genes. Of these, VIPR2, UBE3C, and PTPRN2 are three promising candidate genes for the comorbid CD–MDE phenotype. VIPR2, is one of a group of circadian genes; it encodes the VIPR2 receptor. The circadian genes have been implicated in cocaine sensitization (Andretic et al, 1999) and drug dependence (Perreau-Lenz et al, 2007) as well as mood disorders (Mendlewicz, 2009).
A recent GWAS of response to the antidepressant citalopram in major depressive disorder showed the most significant associated SNP (rs6966038, p=4.65 × 10−7) to be located 51 kb from UBE3C (Garriock et al, 2010). UBE3C encodes E3 ubiquitin ligase, which targets and degrades unneeded or damaged proteins by ubiquitinization and proteolysis; ubiquitin-mediated proteolysis has important roles in various types of substance dependence (Self et al, 1998; French et al, 2001). Taken together, ubiquitin-mediated proteolysis may be important in the development of MDE and drug dependence.
PTPRN2, encodes a member of the protein tyrosine phosphatase (PTP) family. PTPRN2 knockout mice showed decreased insulin secretion and significantly decreased release of norepinephrine, dopamine and 5-HT function in brain, which led to changes in anxiety-like behavior and learning (Nishimura et al, 2009).
In a previous linkage study of comorbid alcoholism and depression, Nurnberger et al (2001) reported a lod score of 3.97 for depression on chromosome 7 at 150 cM in a discovery data set that was predominantly EA subjects, but not in a replication data set. That locus is approximately 33 cM from our linkage peak for MDE and CD–MDE in the AA sample.
The linkage region on chromosome 10 for CD, CD–MDE, and AD in our previous report, contains at least three candidate genes whose protein products are involved in G protein-coupled receptor (GPCR) signaling: GRK5 (GPCR kinase 5), RGS10 (regulator of G protein signaling 10), and GPR26 (GPCR 26).
As all the families in this study were ascertained through affected sib-pairs for CD or OD, there are very few subjects having MDE without a substance dependence disorder. Thus, it is difficult to discern whether linkage findings for the CD–MDE phenotype represent a locus for a subtype of CD or for MDE alone. The linkage peaks on chromosomes 2 and 7 in the AA families and on chromosome 16 in the EA families (with similar linkage peaks for CD–MDE and MDE, but almost none for CD only), suggest the existence of a CD subtype (CD comorbid with MDE). However, the non-overlapping linkage peaks in the EA families on chromosome 13 for MDE alone and on chromosomes 5 and 10 for CD–MDE favor the alternative hypothesis: namely, that the findings are best explained by a gene predisposing to MDE. These two linkage signal patterns for CD, MDE, and CD–MDE might co-exist, because the complexity of comorbid addictive (eg, CD) and psychiatric (eg, depression) disorders might involve shared genetic liability with individual-specific genes mediating the development of different disorders. Future studies are needed to identify the gene(s) associated with the CD–MDE phenotype to clarify its relationship with the constituent phenotypes of CD and MDE.
Depression phenotypes are quite heterogeneous and include those that are recurrent and early onset, comorbid with anxiety disorders or bipolar disorders and so on. The most replicated linkage regions associated with depression (any subtype) include chromosomes 3 (105 cM), 8 (25.1 cM), 12 (100–105 cM), 15 (105–115 cM), 17 (28.0 cM), and 18 (75–88 cM) (Abkevich et al, 2003; Camp et al, 2005; Holmans et al, 2004, 2007; Levinson et al, 2007). The linkage regions reported for CD and related phenotypes include regions of chromosomes 3, 10, 12, and 18 (Gelernter et al, 2005). The linkage signals identified in this study do not coincide with any of the previously identified regions. The different phenotypes examined in previous studies and those examined in this study provide a likely explanation for the distinct linkage regions reported.
The results obtained here differed for the EA and AA samples. Considering that the SNP markers used in the linkage analyses were not identical between the AA and EA samples because of the cleaning procedure and analysis routines that excluded different markers for the two populations, we re-ran the analysis using the largest common set of cleaned SNP markers. The resultant linkage signals remained the same within each sample, excluding the possibility that the population-specific findings were due to the use of different marker sets. Many studies have yielded different linkage signals for the same phenotype in different populations (eg, type 2 diabetes mellitus (Malhotra et al, 2009), cardiovascular-related phenotype (Lynch et al, 2005), schizophrenia (Suarez et al, 2006), and nicotine dependence (Li et al, 2008) and so on). There are several possible mechanisms that may underlie the observation of distinct linkage regions identified between populations, including genetic heterogeneity arising from random genetic drift or differences in adaption, environmental differences, differences in allele frequencies and population history, and random effects pertaining to sampling. For instance, differences in allele frequencies across populations may very well lead to differences in the ability to detect IBD patterns such that the detected linkage signals are different across populations. In view of this, our results appear to indicate that distinct genetic susceptibility regions underlie comorbid CD–MDE in AAs and EAs were detected because of random effects pertaining to the factors mentioned above.
Strengths of our study include the fact that we ascertained >700 families with ASPs for CD and/or OD in AAs and EAs, and the corroboration of most non-parametric linkage findings by the parametric linkage analysis. The veracity of the linkage on chromosomes 5 and 7 is reinforced by the two analytical approaches. One set of results strengthens the other and reduces the possibility of a false positive.
Major depression and CD are common disorders that exact a heavy toll on individuals, families, and society. Clinically, these two disorders affect each other in terms of treatment outcome, the presence of one disorder resulting in poorer treatment outcomes, and higher relapse rates in the other (Hasin et al, 2002; Nunes and Levin, 2004). Subjects are more prone to relapse if either one of the disorders is untreated. In this study, we show that these comorbid disorders share genetic vulnerability. Our GWLS identified several novel chromosomal regions likely to harbor genes for CD–MDE or CD alone. Genes on other chromosomes may also affect this vulnerability. Further studies by genomewide association, pathway analysis, or next generation sequencing are needed to find causative variants in the linkage regions. Elucidating the genetic basis of CD comorbid with MDE to increase our understanding of the etiology of this specific subtype of CD could foster prevention efforts and the development of more effective treatments.
References
Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002). Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30: 97–101.
Abkevich V, Camp NJ, Hensel CH, Neff CD, Russell DL, Hughes DC et al (2003). Predisposition locus for major depression at chromosome 12q22–12q23.2. Am J Hum Genet 73: 1271–1281.
Agís-Balboa RC, Pinna G, Zhubi A, Maloku E, Veldic M, Costa E et al (2006). Characterization of brain neurons that express enzymes mediating neurosteroid biosynthesis. Proc Natl Acad Sci USA 103: 14602–14607.
Andretic R, Chaney S, Hirsh J (1999). Requirement of circadian genes for cocaine sensitization in Drosophila. Science 285: 1066–1068.
Camp NJ, Lowry MR, Richards RL, Plenk AM, Carter C, Hensel CH et al (2005). Genome-wide linkage analyses of extended Utah pedigrees identifies loci that influence recurrent, early-onset major depression and anxiety disorders. Am J Med Genet B Neuropsychiatr Genet 135: 85–93.
Davis LL, Frazier E, Husain MM, Warden D, Trivedi M, Fava M et al (2006). Substance use disorder comorbidity in major depressive disorder: a confirmatory analysis of the STAR*D cohort. Am J Addict 15: 278–285.
Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH et al (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11: 446–450.
Eser D, Romeo E, Baghai TC, di Michele F, Schüle C, Pasini A et al (2006). Neuroactive steroids as modulators of depression and anxiety. Neuroscience 138: 1041–1048.
Evans DL, Charney DS (2003). Mood disorders and medical illness: a major public health problem. Biol Psychiatry 54: 177–180.
French SW, Mayer RJ, Bardag-Gorce F, Ingelman-Sundberg M, Rouach H, Neve And E (2001). The ubiquitin-proteasome 26s pathway in liver cell protein turnover: effect of ethanol and drugs. Alcohol Clin Exp Res 25 (5 Suppl ISBRA): 225S–229S.
Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD et al (2010). A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry 67: 133–138.
Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J et al (2009). Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry 65: 111–115.
Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J et al (2006). Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet 78: 759–769.
Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B et al (2005). Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet 5: 45–52.
Goldman D, Oroszi G, Ducci F (2005). The genetics of addictions: uncovering the genes. Nat Rev Genet 6: 521–532.
Hasin D, Liu X, Nunes E, McCloud S, Samet S, Endicott J (2002). Effects of major depression on remission and relapse of substance dependence. Arch Gen Psychiatry 59: 375–380.
Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, Depaulo Jr JR et al (2007). Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am J Psychiatry 164: 248–258.
Holmans P, Zubenko GS, Crowe RR, DePaulo Jr JR, Scheftner WA, Weissman MM et al (2004). Genomewide significant linkage to recurrent, early onset major depressive disorder on chromosome 15q. Am J Hum Genet 74: 1154–1167.
Ingraham LJ, Wender PH (1992). Risk for affective disorder and alcohol and other drug abuse in the relatives of affectively ill adoptees. J Affect Disord 26: 45–51.
Kong A, Cox NJ (1997). Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61: 1179–1188.
Kumar S, Fleming RL, Morrow AL (2004). Ethanol regulation of gamma-aminobutyric acid A receptors: genomic and nongenomic mechanisms. Pharmacol Ther 101: 211–226.
Lander E, Kruglyak L (1995). Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11: 241–247.
Levinson DF, Evgrafov OV, Knowles JA, Potash JB, Weissman MM, Scheftner WA et al (2007). Genetics of recurrent early-onset major depression (GenRED): significant linkage on chromosome 15q25-q26 after fine mapping with single nucleotide polymorphism markers. Am J Psychiatry 164: 259–264.
Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT et al (2008). Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry 13: 407–416.
Lynch AI, Arnett DK, Atwood LD, Devereux RB, Kitzman DW, Hopkins PN (2005). A genome scan for linkage with aortic root diameter in hypertensive African Americans and whites in the Hypertension Genetic Epidemiology Network (HyperGEN) study. Am J Hypertens 18 (5 part 1): 627–632.
MacKenzie EM, Odontiadis J, Le Mellédo JM, Prior TI, Baker GB (2007). The relevance of neuroactive steroids in schizophrenia, depression, and anxiety disorders. Cell Mol Neurobiol 27: 541–574.
Maher BS, Marazita ML, Zubenko WN, Kaplan BB, Zubenko GS (2002). Genetic segregation analysis of alcohol and other substance-use disorders in families with recurrent, early-onset major depression. Am J Drug Alcohol Abuse 28: 711–731.
Malhotra A, Igo Jr RP, Thameem F, Kao WH, Abboud HE, Adler SG et al (2009). Genome-wide linkage scans for type 2 diabetes mellitus in four ethnically diverse populations-significant evidence for linkage on chromosome 4q in African Americans: the Family Investigation of Nephropathy and Diabetes Research Group. Diabetes Metab Res Rev 25: 740–747.
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al (2009). Finding the missing heritability of complex diseases. Nature 461: 747–753.
McMillan I, Robertson A (1974). The power of methods for the detection of major genes affecting quantitative characters. Heredity 32: 349–356.
McPeek MS, Sun L (2000). Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 66: 1076–1094.
Mendlewicz J (2009). Disruption of the circadian timing systems: molecular mechanisms in mood disorders. CNS Drugs 23 (Suppl 2): 15–26.
Ng MY, Levinson DF, Faraone SV, Suarez BK, DeLisi LE, Arinami T et al (2009). Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry 14: 774–785.
Nishimura T, Kubosaki A, Ito Y, Notkins AL (2009). Disturbances in the secretion of neurotransmitters in IA-2/IA-2beta null mice: changes in behavior, learning and lifespan. Neuroscience 159: 427–437.
Nunes EV, Levin FR (2004). Treatment of depression in patients with alcohol or other drug dependence: a meta-analysis. JAMA 291: 1887–1896.
Nurnberger Jr JI, Foroud T, Flury L, Su J, Meyer ET, Hu K et al (2001). Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry 158: 718–724.
O’Connell JR, Weeks DE (1998). PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63: 259–266.
Panhuysen CI, Kranzler HR, Yu Y, Weiss RD, Brady K, Poling J et al (2010). Confirmation and generalization of an alcohol-dependence locus on chromosome 10q. Neuropsychopharmacology 35: 1325–1332.
Perreau-Lenz S, Zghoul T, Spanagel R (2007). Clock genes running amok. Clock genes and their role in drug addiction and depression. EMBO Rep 8 Spec. No: S20–S23.
Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A et al (2005). Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA). Drug Alcohol Depend 80: 303–312.
Prescott CA, Aggen SH, Kendler KS (2000). Sex-specific genetic influences on the comorbidity of alcoholism and major depression in a population-based sample of US twins. Arch Gen Psychiatry 57: 803–811.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575.
Reddy DS (2003). Is there a physiological role for the neurosteroid THDOC in stress-sensitive conditions? Trends Pharmacol Sci 24: 103–106.
Regier DA, Farmer ME, Rae DS, Locke BZ, Keith SJ, Judd LL (1990). Comorbidity of mental disorders with alcohol and other drug abuse. Results from the Epidemiologic Catchment Area (ECA) Study. Jama 264: 2511–2518.
Riolo SA, Nguyen TA, Greden JF, King CA (2005). Prevalence of depression by race/ethnicity: findings from the National Health and Nutrition Examination Survey III. Am J Public Health 95: 998–1000.
SAMSHA (2009). Results from the 2008 National Survey on Drug Use and Health: National Findings Office of Applied Studies NSDUH Series H-36. SAMSHA: Rockville, MD.
Self DW, Genova LM, Hope BT, Barnhart WJ, Spencer JJ, Nestler EJ (1998). Involvement of cAMP-dependent protein kinase in the nucleus accumbens in cocaine self-administration and relapse of cocaine-seeking behavior. J Neurosci 18: 1848–1859.
Suarez BK, Duan J, Sanders AR, Hinrichs AL, Jin CH, Hou C et al (2006). Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample. Am J Hum Genet 78: 315–333.
Tanchuck MA, Long SL, Ford MM, Hashimoto J, Crabbe JC, Roselli CE et al (2009). Selected line difference in the effects of ethanol dependence and withdrawal on allopregnanolone levels and 5alpha-reductase enzyme activity and expression. Alcohol Clin Exp Res 33: 2077–2087.
Uhl GR, Grow RW (2004). The burden of complex genetics in brain disorders. Arch Gen Psychiatry 61: 223–229.
Acknowledgements
We are grateful to the volunteer families and individuals who participated in this research study. Genotyping services were provided by the Center for Inherited Disease Research (CIDR) and Yale (Keck Center). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). Genotyping was supported in part by a Yale CTSA and NIH Neuroscience Microarray Consortium award U24 NS051869-02S1. We thank Dr Joan Kaufman at for reading the paper and giving insightful comments. We are grateful to Ann Marie Lacobelle and Greg Kay for their excellent technical assistance. This study was supported by National Institute on Drug Abuse (NIDA) Grants K01 DA24758, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and the VA Connecticut REAP center, a VA MERIT grant, and the VA Connecticut MIRECC Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Drs Yang, Han, and Gelernter report no financial relationships with commercial interests. Dr Gelernter reports that he has received compensation for professional services in the previous 3 years from the following entities: Yale University School of Medicine, Veterans Affairs Healthcare System (VA), and the National Institutes of Health (NIAAA, NIDA, and NIMH) and related to academic lectures and editorial functions in various scientific venues (including the ACNP). Dr Kranzler reports consulting arrangements with Alkermes, GlaxoSmithKline, and Gilead Sciences, and has received research support from Merck and Company, Bristol-Myers Squibb Company. He also reports associations with Eli Lilly, Janssen, Schering Plough, Lundbeck, Alkermes, GlaxoSmithKline, Abbott, and Johnson and Johnson, as these companies provide support to the ACNP Alcohol Clinical Trials Initiative (ACTIVE) and Dr Kranzler receives support from ACTIVE. Dr Farrer received a research grant from Eisai Pharmaceuticals and consultant fees from Novartis Pharmaceuticals.
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, BZ., Han, S., Kranzler, H. et al. A Genomewide Linkage Scan of Cocaine Dependence and Major Depressive Episode in Two Populations. Neuropsychopharmacol 36, 2422–2430 (2011). https://doi.org/10.1038/npp.2011.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2011.122
Keywords
This article is cited by
-
Variants near CHRNB3-CHRNA6 are associated with DSM-5 cocaine use disorder: evidence for pleiotropy
Scientific Reports (2014)