Abstract
The expression of the galanin gene (GAL) in the paraventricular nucleus (PVN) and in the amygdala of higher vertebrates suggests the requirement for highly conserved, but unidentified, regulatory sequences that are critical to allow the galanin gene to control alcohol and fat intake and modulate mood. We used comparative genomics to identify a highly conserved sequence that lay 42 kb 5′ of the human GAL transcriptional start site that we called GAL5.1. GAL5.1 activated promoter activity in neurones of the PVN, arcuate nucleus and amygdala that also expressed the galanin peptide. Analysis in neuroblastoma cells demonstrated that GAL5.1 acted as an enhancer of promoter activity after PKC activation. GAL5.1 contained two polymorphisms; rs2513280(C/G) and rs2513281(A/G), that occurred in two allelic combinations (GG or CA) where the dominant GG alelle occurred in 70-83 % of the human population. Intriguingly, both SNPs were found to be in LD (R2 of 0.687) with another SNP (rs2156464) previously associated with major depressive disorder (MDD). Recreation of these alleles in reporter constructs and subsequent magnetofection into primary rat hypothalamic neurones showed that the CA allele was 40 % less active than the GG allele. This is consistent with the hypothesis that the weaker allele may affect food and alcohol preference. The linkage of the SNPs analysed in this study with a SNP previously associated with MDD together with the functioning of GAL5.1 as a PVN and amygdala specific enhancer represent a significant advance in our ability to understand alcoholism, obesity and major depressive disorder.
Similar content being viewed by others
INTRODUCTION
The neuropeptide, galanin (GAL), is a 30 amino acid neuropeptide that is expressed in a number of discrete regions of the brain that include the paraventricular nucleus (PVN), the arcuate nucleus of the hypothalamus, and the amygdala (Ceccatelli et al, 1992; Cortes et al, 1990; Kaplan et al, 1988; Miller et al, 1993a; Planas et al, 1994b).
Expression of GAL in the PVN has been shown to positively influence fat intake and preference in rodents (Adams et al, 2008; Karatayev et al, 2009a, 2009b; Leibowitz, 2005, 2007; Leibowitz and Kim, 1992; Poritsanos et al, 2009). Furthermore, recent studies in humans have shown the presence of polymorphisms strongly associated with elevated triglyceride levels in the blood (Plaisier et al, 2009).
Expression of GAL in the PVN also modulates ingestion of ethanol (Karatayev et al, 2009b, 2009c; Leibowitz et al, 2003) and a possible involvement of the GAL gene in susceptibility to alcoholism was suggested by an association study carried out on Finnish and Plains-Indian populations (Belfer et al, 2006). Links between the expression of GAL in the hypothalamus and modulation of the drug reward system of the mesolimbic dopaminergic system have prompted an interest in GAL and addictive behaviors (Picciotto et al, 2010). Although the mechanisms are unclear, injection of GAL into the PVN increases dopamine release from the nucleus accumbens, which is the center of the brain responsible for addictive behavior (Rada et al, 1998). Thus, it is possible that mis-expression of GAL in the PVN may contribute to the rewarding effects of alcohol.
Targeted deletions of the GAL gene and pharmacological manipulation of its receptors have also been shown to influence anxiety and mood-related behavior in mice (Barr et al, 2006; Holmes et al, 2003; Holmes and Picciotto, 2006; Kuteeva et al, 2007, 2008a, 2008b). Furthermore, polymorphisms in and around the GAL gene have been associated with panic disorder and anxiety in female patients (Unschuld et al, 2008, 2010). As a result of its expression in the amygdala (Miller et al, 1993b; Planas et al, 1994a) and its known role in mood modulation (Crawley et al, 2002; Hobson et al, 2006; Karlsson and Holmes, 2006; Madaan and Wilson, 2009; Paschos et al, 2009; Rotzinger et al, 2009), GAL and its three receptors have received a great deal of attention by researchers attempting to understand the role of the galaninergic system in chronic anxiety and depression and to develop novel therapies. Indeed, a recent GWAS analysis demonstrated evidence for an association between polymorphisms close to the GAL gene locus and major depressive disorder (Wray et al, 2010).
Identifying and characterizing the regulatory sequences responsible for delimiting the expression in the PVN and amygdala, and how these sequences may have been altered by human variation, is a major scientific priority for those interested in the role of GAL in the control of alcohol intake, fat intake, and mood modulation. For example, one SNP (rs948854) was discovered 2 kb from the GAL locus and was associated with panic disorder in women (Unschuld et al, 2008, 2010). However, what effects this polymorphism had on the activity of the GAL promoter was not reported. Further analysis of a 20 kb region flanking the GAL gene succeeded in identifying a regulatory region capable of being induced following sensory nerve axotomy. However, no activity was reported in any other region of the brain associated with the expression of the GAL gene (Bacon et al, 2007; Rokaeus and Waschek, 1998). These studies suggest that the sequences essential to PVN and amygdala-specific expression of the GAL gene lie >20 kb away from the GAL transcriptional start site.
The very precise expression pattern of the GAL gene seen in the PVN in a number of different species including rats, mice (Ceccatelli et al, 1989), sheep (Barker-Gibb and Clarke, 1996), and birds (Azumaya and Tsutsui, 1996) suggests that the regulatory systems responsible are critical for the proper function of GAL and have not changed for hundreds of millions of years. We therefore used comparative genomics and transgenic analysis to explore the location of remote, polymorphic, and highly conserved, regulatory regions that may be responsible for driving the expression of the GAL gene in these regions of the brain. We also used transfection of primary hypothalamic neuron cultures with reporter genes to determine whether common allelic variations of these enhancers showed significant differences in their activities in these cells. Considering the important roles played by GAL gene expression in the PVN and the amygdala these studies may provide a unique opportunity to understand the cellular systems that control GAL regulated appetite, mood and alcohol intake and how human variation in these systems may change their activity and contribute to obesity, major depressive disorder and alcoholism.
MATERIALS AND METHODS
Bioinformatics and Sequence Analysis
Evolutionary conservation was identified by genomic comparison using the ECR Browser (Ovcharenko et al, 2004) and the UCSC genome browser (Kent et al, 2002). Prediction of the effects of SNPs on transcription factor—DNA binding was carried out using the newly developed RegSNP (http://viis.abdn.ac.uk/regsnp/Home.aspx) (Davidson et al, submitted). SNP linkage analysis was carried out on the UCSC genome browser.
Plasmid Construction
The GAL5.1 region was amplified from human placental DNA using a high fidelity polymerase (Roche, Expand HI FI kit), and the following oligonucleotide primers: GAL5.1_F: 5′-TGCTCCTGGATGGAGAGAAG-3′, GAL5.1_R: 5′-CTCCATCAGAGACGCCAAAC-3′. After confirmation of correct amplification by DNA sequencing, the PCR product was restriction digested with ApaI and SspI and cloned into pGEM-5Z (Promega) that had been restriction digested with ApaI and EcoRV to produce pGAL5.1GEM5. The GAL5.1-hβg-lacZ construct containing the human beta-globin promoter (hβg) and the β-galactosidase reporter gene (LacZ) was created by removing the GAL5.1 element from the pGAL5.1GEM5 plasmid and inserting it into the p1230 plasmid (a kind gift from Robert Krumlauf) by restriction digest with SalI and ApaI. Allelic variants of the GAL5.1 region were created in pGAL5.1GEM5 by site directed mutagenesis using a Quick Change II kit (Stratagene) using the following primer pairs: rs2513280 forward primer 5′-GTGGTAATTAAAGTAATGTCCTGTGCTCAAATTGCTTGTGC-3′, reverse primer 5′-GCACAAGCAATTTGAGCACAGGACATTACTTTAATTACCAC-3′; rs2513281 forward primer 5′-ATGACTGTGTAACGTTCGTCACCTTTATTTTTGTATCTGTAAATGC-3′, reverse primer 5′-GCATTTACAGATACAAAAATAAAGGTGACGAACGTTACACAGTCAT-3′. Luciferase constructs were created by inserting the different allelic variants of the GAL5.1 element from pGAL5.1GEM5 into the SmaI and NotI restriction sites of the pTAL-Luc plasmid (Clonetech, abbreviated to pLuc) to form pGAL5.1(GG)-Luc, pGAL5.1(GA)-Luc, pGAL5.1(CG)-Luc, and pGAL5.1(CA)-Luc (Figure 3a).
Transgenic Mouse Production
Following linearization and removal of the plasmid backbone, the GAL5.1-hβg-lacZ construct DNA was microinjected into 1-cell C57/BL6xCBA F1 mouse embryos at a concentration of 2–4 ng/μl as previously described (Nagy et al, 2003). Surviving embryos were oviduct transferred into pseudopregnant CD1 host mothers.
Analysis of LacZ Gene Expression in Transgenic Lines
Six-week-old F1 transgenic mice from lines 18 and 21 were humanely killed by lethal injection of euthatol according to current UK Home Office guidelines. Following 4 h of fixation in 4% paraformaldehyde in phosphate-buffered saline (PFA-PBS), brain sections were then stained using 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) solution for 4–12 h as previously described (Nagy et al, 2003). X-gal stained tissues were then prepared for vibratome sectioning as previously described (Davidson et al, 2006b). 50-μm sections were then cut on a Vibratome series 1000. Sections were mounted on glass slides and photographed under light field illumination.
In situ Hybridization
Radioactive in situ hybridization was carried out on 10-μm thick sections of 6-week-old mouse brain as previously described (Mercer et al, 1996).
Immunofluorescence
Brain tissues derived from 6-week-old GAL5.1-hβg-LacZ transgenic mice (line 21) were fixed in 4% PFA-PBS for 4 h and prepared for microtome sectioning by allowing tissue to equilibrate in 30% sucrose in PBS at 4 °C. In all, 60-μm sections were then cut on a Microtome (American Optical Company) and maintained as free floating sections within Netwells (Sigma) in a 12-well plate. Free floating sections were washed with antibody buffer (PBS containing 0.1% Triton-X100 and 0.3 M NaCl) and allowed to incubate overnight at 4 °C with rabbit anti-GAL 1/1000 (Millipore, Peninsula Labs) and chicken anti-β-galactosidase 1/300 (Abcam) primary antibodies diluted in antibody buffer. Secondary antibody used were donkey anti-rabbit (Invitrogen, Alexa 488) and donkey anti-chicken (Invitrogen, Alexa 594) in antibody buffer overnight at 4 °C. Sections were then mounted on glass slides with and sealed with VECTASHIELD mounting medium with DAPI (Vector labs). Immunofluorescence was visualized and images of 1-m optical sections obtained using a Zeiss 510 laser scanning confocal microscope.
Cell Culture and Luciferase Reporter Gene Assays
In order to recover sufficient cells, the whole hypothalamus region, as previously defined (Paxinos, 1997), was dissected from 1- to 3-day-old rat neonates into Neurobasal-A medium (Invitrogen) and treated with 0.05% trypsin/EDTA for 15 min. Trypsinization was stopped by the addition of trypsin inhibitor followed by replacement into fresh Neurobasal-A medium. Tissues were triturated with a fire polished Pasteur pipette before being filtered through a cell strainer to create a single cell suspension that was cultured in Neurobasal-A medium with B27 additive and glutamax (Invitrogen). These cells were transfected with plasmid DNA using Neuromag according to the manufacturer's instructions (OZ Bioscience). Briefly, Neuromag reagent was added to a plasmid-media preparation and incubated at room temperature for 15 min allowing 250 ng of plasmid DNA in the media per 150 000 cells to be transfected. The DNA-Media-Neuromag solution was then added to previously prepared single cell cultures in 24-well plates and incubated on a magnetic plate for 15 min in a cell culture incubator at 37 °C and 5% CO2.
SHSY-5Y neuroblastoma cells were cultured as previously described (Gillies et al, 2010) and magnetofected as described above.
Luciferase activity of plasmid constructs were measured using dual luciferase assay kits (Promega) using lysates from cell cultures transfected with the GAL5.1 luciferase constructs. Cell cultures were lysed and the proteins stabilized using the passive lysis buffer as per the manufacturer's instructions (Promega). The dual luciferase assay analysis was carried out on a glomax 96-microplate luminometer (Promega) using 20 μl of cell lysate per well of a white 96-well plate.
RESULTS
Identification, Isolation, and Cloning of GAL5.1
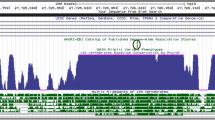
We have previously shown that comparative genomic analysis is an effective method of detecting functional gene regulatory regions (Davidson et al, 2006a, 2006b; Mackenzie et al, 1997, 2004a; MacKenzie and Quinn, 2004b; Miller et al, 2007, 2008; Shanley et al, 2010). We carried out comparative analysis of 100 kb of the genome surrounding the human GAL locus (Figures 1a and b) using the monkey, dog, mouse, rat, opossum, and chicken genomes and succeeded in identifying an area of homology that was located 42 kb 5′ of the GAL transcriptional start site that we called GAL5.1. This sequence had been conserved since bird–mammal divergence reflecting 310 million years of conservation.
(a) VISTA plot from the ECR Browser comparing 45 kb of DNA to the 5′ of the GAL locus with the genomes of (from top to bottom) chicken, marsupial, mouse, dog, and rhesus monkey. The x axis represents linear distance with reference to the human genome sequence. The y axis represents levels of sequence conservation between 50 and 100%. Blue lines with chevrons represent the genomic extent of each gene. Red, green, blue, pink, and yellow peaks represent areas of sequence conservation (>75% over 100 bp) in intergenic non-coding, repetitive sequence, exonic, intronic, and untranslated regions respectively. (b) A vertebrate Multiz-alignment and phast-con 28 species conservation plot showing where the rs2513280 and rs2513281 SNPs fall in relation to the most conserved areas of the enhancer sequence. (c, d) Multiple alignment plots of human, chimp, resus monkey, mouse rat, and chicken DNA within the GAL5.1 enhancer and surrounding the (c) rs2513280 and (d) rs2513281 loci demonstrating levels of conservation (red boxes) and aligned binding matrices of a number of different transcription factor proteins that are known to be expressed within the hypothalamus. The tables above both alignments (c, d) display predicted probabilities of transcription factor binding based on the known binding matrix of each transcription factor where 1=perfect match and 0=no match as predicted by the regSNP algorithm. Thus, column 1 represents binding probabilities to the dominant G allele for both loci and column 2 represent those of the (c) C and (d) A alleles. The color reproduction of this figure is available at the Neuropsychopharmacology journal online.
Gal5.1 Supports Gene Expression in the Amygdala and Hypothalamus in Regions that also Express GAL
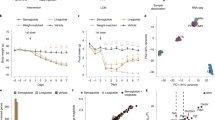
We used high fidelity PCR to isolate the GAL5.1 sequence and cloned it into a LacZ reporter plasmid driven by the previously characterized hβ-globin promoter (Yee and Rigby, 1993) to form GAL5.1-hβg-LacZ. Two transgenic mouse lines (18 and 21) were generated using the GAL5.1-hβg-LacZ construct (see Figures 2a, b and 3a) and both lines were analyzed in parallel for the expression of the β-galactosidase (LacZ) marker. Brain and spinal cord were dissected from individuals from each line and incubated overnight in X-gal stain. We also examined the expression of the endogenous GAL gene using radioactive in situ hybridization and noticed a remarkable correlation in the expression of the transgene with the distribution of GAL mRNA (Figures 2a–c and e–g). In the hypothalamus, there was strong expression of β-gal in areas, which included the PVN, the ventromedial hypothalamic nucleus and the arcuate nucleus in both of the GAL5.1-LacZ transgenic lines generated (Figures 2a–c and e–g). In addition, we observed activity of both the transgene and the GAL gene within the tuberal nucleus (Figures 2e–g) as previously reported (Evans et al, 1993). Expression was also seen in the medial and central amygdala (Figures 2a–c and e–g). These observations were confirmed at the cellular level using fluorescent immunohistochemistry on 10-μm sections through the amygdala and hypothalamus of GAL5.1-hβg-LacZ transgenic mouse using antibodies against GAL and β-galactosidase that were analyzed using 1-μm optical sections using a confocal microscope (Figures 2d and h). Although there were a number of non-neuronal areas of transgene activity in areas such as the vasculature of the pancreas this expression was not consistently observed in both lines. This was also true of expression of LacZ observed in a subpopulation of cells in the dorsal root ganglia where GAL is known to be expressed in sensory neurons. However, this expression was only observed in a few individuals of one line (21) so may have represented an insertional effect.
(a, b, e, f) Photomicrographs of the expression of the β-galactosidase reporter gene, as detected using X-gal staining, within the (a, b) the hypothalamic region and (e, f) the amygdala and arcuate nucleus region of transgenic lines (a) 21 and (b) 18 of mice containing the GAL5.1hβg-LacZ reporter construct. (c, g) Expression of GAL mRNA within the (c) hypothalamus and (g) amygdala region of the mouse brain by in situ hybridization. (d, h) In all, 1-μm optical confocal sections through the (d) paraventricular nucleus and (h) amygdala region of a mouse transgenic for the GAL5.1hβg-lacZ transgene (line 21) following fluorescent immunohistochemistry using antisera against the GAL peptide (false color green) and β-galactosidase (false color red). Colocalization within cells is indicated by false color yellow and highlighted with white arrows. Nucleii are stained with TO-PRO3 (blue). Arc, arcuate nucleus; CeA, central amygdala; MeA, medial amygdala; PVN, paraventricular nucleus; TU, tuberal nucleus; VMH, ventromedial hypothalamic nucleus. Scale bar in (d) and (h)=20 μm.
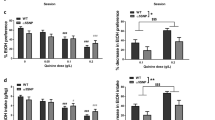
(a) Diagrammatical representation of the different constructs used in the current study (not to scale) and showing the different alleles reproduced in each construct using site directed mutagenesis. LacZ, β-galactosidase reporter, hβgprom, human β-globin promoter, TKprom, herpes simplex virus thymidine kinase promoter. (b) Graph comparing levels of induction of the pLuc plasmid against that of the pGAL5.1(GG)-Luc plasmid (Figure 3a) following their transfection into SHSY-5Y cells and treatment with forskolin, angiotensin II, or PMA. Relative levels of induction are normalized against renilla luciferase and vehicle control (DMSO or H2O). (c) Graph showing the relative expression levels of the luciferase reporter constructs shown in Figure 3a and normalized to co-transformed renilla expressing plasmid and the pLuc plasmid. *p<0.05; **p<0.01; ns, not significant.
Gal5.1 can Enhance Promoter Activity Following Activation of PKA Pathways
We next sought to determine whether the GAL5.1 sequence could act as an enhancer of promoter activity thus acting as a classical enhancer sequence. In addition, we further explored the identity of the cellular transduction systems that influenced the activity of the GAL5.1 sequence. We transfected SHSY-5Y neuroblastoma cells (Scott et al, 1986) with either the pTAL-Luc vector (abbreviated to pLuc) or pGAL5.1(GG)-Luc plasmid and cultured these cells in the absence or presence of agonists of the PKA (forskolin), PKC (phorbol ester PMA), or MAPkinase (angiotensin II) pathways. None of the agonists used had any effect on the herpes simplex virus thymidine kinase (HSV-TK) promoter contained within pLuc (Figure 3b). In addition, in the absence of any agonists the GAL5.1 sequence also demonstrated no significant ability to further enhance promoter activity in these cells (Figure 3b). Moreover, the use of angiotensin II or forskolin also had no significant effect. However, addition of PMA induced a significant enhancement in the ability of GAL5.1 to induce promoter activity (Figure 3b). These studies suggest that GAL5.1 is an enhancer sequence that responds to activation of the PKC signalling cascade.
Bioinformatic Analysis of Allelic Variants of GAL5.1
As a result of the possibility that GAL mis-regulation may have a role in obesity, alcoholism, or mood disorders, we explored whether there were polymorphisms of the GAL5.1 enhancer in the human population that might alter its activity. Using the dbSNP database and the UCSC browser we identified two human polymorphisms within the GAL5.1 enhancer, rs2513280 (G/C) and rs2513281 (G/A) that occur in GAL5.1 sequence (Figures 1b–d). Multiple species alignments show that in all vertebrate species studied the G allele is conserved at the non-human equivalents of both the rs2513280 and rs2513281 loci (Figures 1c and d). The conservation of a G residue through evolution at both these sites reflects the allelic distribution in the human population whereby 70–83% of alleles, at both these loci, are G residues. However, a significant proportion of the European (16.7%), African (26.2 and 20%), and Asian (29.2%) populations maintain C and an A alleles at the rs2513280 and rs2513281 loci, respectively. Heatmap analysis using Hapmap shows that these alleles are very closely linked (LOD=14.97, D′=1) to the degree that there is little evidence in any population of genotypes other than GG or CA. Intriguingly, rs2156464, which had previously been linked to major depressive disorder (Wray et al, 2010) was found to be in LD with rs2513280 and rs2513281 with an R2 of 0.687. However, significant linkage disequilibrium with other SNPs in and around the galanin locus, that have been previously associated with depressive illness, food intake and alcoholism, such as rs948854 and rs4432027 (Unschuld et al, 2008, 2010) rs694066 (Ruano et al, 2006) and rs3136540 (Belfer et al, 2006) was not detected.
In order to predict the effects of these polymorphisms on the activity of the GAL5.1 enhancer we developed a novel algorithm, called RegSNP that predicts the effects of human polymorphisms on the binding sites of all the know transcription factor-binding matrices contained within the Transfac database (Davidson et al, submitted). RegSNP holds the binding matrices of all of the transcription factors contained in the Transfac database. Using regSNP, we were able to predict the ability of a number of transcription factors, known to be expressed in the hypothalamus, to bind the CA allele. In the case of rs2513280, the predicted decrease in the binding of c-ETS-1 was interesting as c-ETS-1 is expressed strongly in the PVN (Figure 2a inset from Allen brain atlas). In rs2513281, the change from a G to an A residue decreased the predicted affinity of the thyroid receptor (T3R), PXR, TCF, and WT1. However, the A allele at this locus was predicted to increase the ability to bind AP-1 and the Kruppel-like transcription factors.
The Dominant GG Allele of GAL5.1 Demonstrates Significantly Higher Activity than the CA Activity in Hypothalamic Neurones
It has been suggested that changes in the regulation of gene expression, because of polymorphic variation, is a major contributing factor in disease susceptibility. As the GAL gene has such an important role in the PVN and the amygdala, we explored whether the polymorphic variants we had discovered affected the activity of the GAL5.1 enhancer in hypothalamic neurones. We used site directed mutagenesis to recreate known human polymorphic variants of the GAL5.1 enhancer and cloned these variants into a luciferase reporter construct (Figure 3a). These constructs were transfected into primary neonate rat hypothalamic neurones together with a renilla luciferase control plasmid using neuromag magnetic transfection technology. After 24 h, cells were lysed and lysates assayed using a dual luciferase assay. Significant differences were observed in the ability of these different polymorphic variants of GAL5.1 to drive luciferase expression in these primary hypothalamic neurones (Figure 3c). The Gal5.1(GG)-luc construct that contains the GG allele, which has been evolutionary conserved and represents the major haplotype in human populations, showed the highest level of luciferase expression. Relative to the Gal5.1(GG)-luc construct luciferase activity from GAL5.1(GA)-Luc was reduced by 60%, while the GAL5.1(CG)-Luc and GAL5.1(CA)-Luc demonstrated a 35 and 40% decrease in activity, respectively (Figure 3c).
DISCUSSION
Although neuropeptides in the hypothalamus have been widely studied in the context of obesity (Lu et al, 2007), alcoholism and to a lesser extent, in the amygdala and mood disorders (Sergeyev et al, 2005), there is little evidence that mutations or polymorphisms in the coding sequences of the genes that encode these neuropeptides have a major role in conferring susceptibility to these disorders. However, evidence suggests that polymorphic variation of regions of the genome that control the expression of genes, at the level of transcription, splicing, or translation, may be major contributory factors in conferring disease susceptibility (Wray, 2007). This was confirmed following the results of a recent meta-analysis of data from multiple GWA studies that concluded that 88% of disease associated ‘hits’ occur within the non-coding genome (Hindorff et al, 2009; Singleton et al, 2010). The high prevalence of disease causing SNPs in the non-coding genome, when combined with the comparative lack of understanding of the non-coding genome in gene regulation and a relative inability to efficiently analyze this uncharted part of the genome has led to much concern. Thus, in addition to determining the mechanisms that control the expression of GAL in the hypothalamus and the amygdala, this study sought to demonstrate the combined use of comparative genomics, transgenic analysis, and primary cell analysis to identify, characterize, and assess the effects of human variation on the regulatory regions that maintain the expression of the GAL gene in these critical parts of the brain.
The high degree of conservation of the patterns of expression of the GAL gene in the hypothalamus and amygdala suggests that the mode of action of GAL depends on its expression in very specific groups of cells. Thus, the evidence suggests that changes in GAL expression in these regions were inconsistent with species survival during evolution. However, it is clear from previous studies that 20 kb of the promoter region of the GAL gene could not produce marker gene expression in the brain of transgenic mice (Bacon et al, 2007). Therefore, before this study, a critical functional component of the GAL locus remained unidentified. By virtue of its extreme conservation, this study succeeded in predicting and identifying a regulatory region that lay 42 kb 5′ of the human GAL gene and was able to support the expression of marker genes in cells of the amygdala, PVN, and arcuate nucleus that also express GAL mRNA. The importance of GAL gene regulation in the mode of action of GAL is highlighted by the extreme conservation of GAL5.1, which greatly exceeds the conservation of the GAL gene coding region itself. This degree of conservation demonstrates that, while there has been a significant degree of sequence change in the GAL coding region since bird–mammal divergence, evolution has maintained strong selection against changes in the GAL5.1 sequences to ensure the correct expression of GAL in these regions of the brain.
It has been shown that cis-regulatory sequences with importance to the expression of specific genes are not only highly conserved but are retained in cis with these genes often for hundreds of millions of years (Mackenzie et al, 2004a; Mongin et al, 2009; Navratilova and Becker, 2009a; Navratilova et al, 2009b). This appears to be the case with the GAL5.1 enhancer and the GAL gene, which have remained in-cis for at least 310 million years in all of the higher vertebrate species examined to date. Considering the propensity of genomes to rearrange through evolution this strong ‘in-cis’ relationship suggests that separation of the GAL gene and GAL5.1 is inconsistent with vertebrate survival and serves as the first piece of evidence functionally linking the GAL5.1 sequence to the GAL gene.
A second piece of evidence is the remarkable similarity of the patterns of expression of GAL5.1-hβg-LacZ transgene derived β-galactosidase and the expression of GAL mRNA in the PVN, the arcuate nucleus and the amygdala of both transgenic lines produced in the current study. The fact that both lines generated were able to produce patterns of expression of the transgene that matched the expression of the endogenous GAL gene argues against the possibility that these patterns were caused by insertional effects. The correspondence in patterns strongly suggests that GAL5.1 is not only involved in GAL expression but that it may be the only cis-regulatory element required for the tissue-specific expression of the GAL gene in the hypothalamus and amygdala.
Third, we also show using high-resolution confocal imaging that the transgene and the endogenous peptide are co-expressed at the cellular level in individual neurones of the amygdala and the PVN.
Together, these three lines of evidence argue strongly in favor of a role for the GAL5.1 enhancer in the regulation of the GAL in the hypothalamus and the amygdala. On the basis of its strong conservation and its activity in GAL-expressing neurones, we predict that genomic deletion of the GAL5.1 element by embryonic stem cell targeting in mice would produce a similar phenotype to that previously observed following deletion of the GAL gene coding region (Wynick and Bacon, 2002).
Although our evidence suggests that GAL5.1 is involved in the regulation of GAL in these tissues we cannot rule out the possibility that specific enhancer–promoter interactions may be required to allow appropriate responses to specific stimuli or signal transduction systems. Thus, it would be interesting to see how GAL5.1 interacted with the GAL promoter regions previously identified (Bacon et al, 2007). Moreover, an interesting contrast can be drawn between GAL5.1 and the ECR2 enhancer shown to control the expression of the TAC1 gene in sensory neurones (Shanley et al, 2010, 2011). ECR2 could only drive tissue-specific expression in the presence of the endogenous TAC1 promoter whereas GAL5.1 can support expression in the presence of an exogenous weak promoter such as the human β-globin promoter or HSV-TK promoter. These observations suggest that different enhancers possess varying levels of autonomy or interdependence and that the GAL5.1 is functionally self-contained relative to some other previously characterized enhancers (Shanley et al, 2010, 2011).
In addition to determining that GAL5.1 is able to drive marker gene expression in cells of the hypothalamus and amygdala that also express the GAL peptide, we wanted to determine quantitatively whether GAL5.1 could enhance the activity of an already well-characterized promoter. We used a human neuroblastoma cell line, called SHSY-5Y, which displays neuronal characteristics (Scott et al, 1986). Using these cells, we were unable to show that GAL5.1 could act as an enhancer of promoter activity. However, we have previously shown that many regulatory sequences are only able to act as enhancers in response to activation of an appropriate signal transduction cascade (Shanley et al, 2010, 2011). Thus, we were able to show that GAL5.1 acted as an enhancer of promoter activity following activation of the PKC pathway, which is known to modulate gene expression in the hypothalamus and amygdala, but not PKA or MAPkinase pathways. These results suggest that GAL5.1 is an enhancer responsible for transmitting the activation of PKC pathways to the core transcriptional apparatus. Intriguingly, it was observed that one of the allelic variants of the GAL5.1 enhancer reduced the predicted ability of the ETS-1 transcription factor to bind. This prediction is interesting as ETS-1 expression overlaps that of the GAL gene and GAL5.1 in the PVN (Figure 2a insert). In addition, a number of studies have shown that the regulatory activity of ETS-1 is modulated by the PKC signal transduction pathway (Naito et al, 2002; Vetter et al, 2005).
In addition to establishing the cells in which the GAL5.1 enhancer is active and that GAL5.1 is activated by PKC activation, we identified two polymorphisms within the GAL5.1 sequence. Examination of the population frequency of these polymorphisms suggests little evidence of the existence of other allele combinations other than GG or CA. In addition, the numbers of individuals homozygous for CA is much lower than expected supporting the hypothesis that the CA allele has reduced fitness in the population in the past. In keeping with this hypothesis, we observed that the equivalent loci within the GAL5.1 enhancer of other species are always GG suggesting that divergence from the GG allele has not been compatible with species fitness during evolution. If we consider our observation that the CA allele is significantly less active in primary hypothalamic neurones than the GG allele, together with the observation that GAL5.1 is active in the PVN and that that GAL expression in PVN controls appetite for fatty foods and alcohol, then it is possible that the CA allele may reduce appetite for these nutrients. Thus, the CA allele may have been selected against through mammalian evolution because maintenance of a preference for high energy fatty foods and alcohol during evolution may have been essential for survival. Moreover, there is little genetic evidence that polymorphisms around the GAL locus are associated with obesity (Schauble et al, 2005). Instead, it may be possible that polymorphisms around the GAL locus, including the CA allele of GAL5.1, might reduce preferences for high fat foods and alcohol. Indeed, one study that examined the effects of different gene polymorphisms on weight loss through reduced carbohydrate intake demonstrated that polymorphisms in the GAL gene were significantly associated with accelerated weight loss (Ruano et al, 2006). Thus, it would be interesting to determine the GAL5.1 allele frequencies in populations of individuals attempting to lose weight to determine if the CA allele can be associated with accelerated weight loss.
With respect to depression and anxiety there have been a number of association studies linking polymorphisms around the GAL locus with mood disorders. For example, one polymorphism, rs948854, was associated with depression and anxiety disorder in female patients together with worse treatment response to antidepressants (Unschuld et al, 2008, 2010). In addition, a larger GWAS study suggested that a polymorphism; rs2156464, that lies 25kb from the GAL locus, was ‘worthy of note’ and rated amongst the top ranked loci associated with major depressive disorder (Wray et al, 2010). Importantly, we observed that rs2156464 was in significant LD with both rs2513280 and rs2513281. This intriguing result is significant considering the role of Galanin in the modulation of mood. Furthermore, the observations that GAL5.1 is active in galanin expressing cells of the amygdala and that allelic variants of GAL5.1 display differential activities suggests a mechanism whereby those variants might alter levels of galanin expression and contribute to depressive disease. Although the association of polymorphisms around the Galanin locus with mood disorders requires replication we believe that there is enough evidence present to warrant further exploration of the GAL5.1 element in the amygdala and the contribution of its allelic variants to depressive disorders.
References
Adams AC, Clapham JC, Wynick D, Speakman JR (2008). Feeding behaviour in galanin knockout mice supports a role of galanin in fat intake and preference. J Neuroendocrinol 20: 199–206.
Azumaya Y, Tsutsui K (1996). Localization of galanin and its binding sites in the quail brain. Brain Res 727: 187–195.
Bacon A, Kerr NC, Holmes FE, Gaston K, Wynick D (2007). Characterization of an enhancer region of the galanin gene that directs expression to the dorsal root ganglion and confers responsiveness to axotomy. J Neurosci 27: 6573–6580.
Barker-Gibb ML, Clarke IJ (1996). Increased galanin and neuropeptide-Y immunoreactivity within the hypothalamus of ovariectomised ewes following a prolonged period of reduced body weight is associated with changes in plasma growth hormone but not gonadotropin levels. Neuroendocrinology 64: 194–207.
Barr AM, Kinney JW, Hill MN, Lu X, Biros S, Rebek Jr J et al (2006). A novel, systemically active, selective galanin receptor type-3 ligand exhibits antidepressant-like activity in preclinical tests. Neurosci Lett 405: 111–115.
Belfer I, Hipp H, McKnight C, Evans C, Buzas B, Bollettino A et al (2006). Association of galanin haplotypes with alcoholism and anxiety in two ethnically distinct populations. Mol Psychiatry 11: 301–311.
Ceccatelli S, Eriksson M, Hokfelt T (1989). Distribution and coexistence of corticotropin-releasing factor-, neurotensin-, enkephalin-, cholecystokinin-, galanin- and vasoactive intestinal polypeptide/peptide histidine isoleucine-like peptides in the parvocellular part of the paraventricular nucleus. Neuroendocrinology 49: 309–323.
Ceccatelli S, Giardino L, Calza L (1992). Response of hypothalamic peptide mRNAs to thyroidectomy. Neuroendocrinology 56: 694–703.
Cortes R, Villar MJ, Verhofstad A, Hokfelt T (1990). Effects of central nervous system lesions on the expression of galanin: a comparative in situ hybridization and immunohistochemical study. Proc Natl Acad Sci USA 87: 7742–7746.
Crawley JN, Mufson EJ, Hohmann JG, Teklemichael D, Steiner RA, Holmberg K et al (2002). Galanin overexpressing transgenic mice. Neuropeptides 36: 145–156.
Davidson S, Miller KA, Dowell A, Gildea A, Mackenzie A (2006a). Cellular co-expression of a LacZ marker gene driven by the amygdala-specific ECR1 enhancer with the substance P neuropeptide. Mol Psychiatry 11: 323.
Davidson S, Miller KA, Dowell A, Gildea A, Mackenzie A (2006b). A remote and highly conserved enhancer supports amygdala specific expression of the gene encoding the anxiogenic neuropeptide substance-P. Mol Psychiatry 11: 410–421.
Evans HF, Huntley GW, Morrison JH, Shine J (1993). Localisation of mRNA encoding the protein precursor of galanin in the monkey hypothalamus and basal forebrain. J Comp Neurol 328: 203–212.
Gillies SG, Haddley K, Vasiliou SA, Jacobson GM, von Mentzer B, Bubb VJ et al (2010). Distinct gene expression profiles directed by the isoforms of the transcription factor neuron-restrictive silencer factor in human SK-N-AS neuroblastoma cells. J Mol Neurosci 44: 77–90.
Hindorff LA, Burke W, Laberge AM, Rice KM, Lumley T, Leppig K et al (2009). Motivating factors for physician ordering of factor V Leiden genetic tests. Arch Intern Med 169: 68–74.
Hobson SA, Holmes FE, Kerr NC, Pope RJ, Wynick D (2006). Mice deficient for galanin receptor 2 have decreased neurite outgrowth from adult sensory neurons and impaired pain-like behaviour. J Neurochem 99: 1000–1010.
Holmes A, Kinney JW, Wrenn CC, Li Q, Yang RJ, Ma L et al (2003). Galanin GAL-R1 receptor null mutant mice display increased anxiety-like behavior specific to the elevated plus-maze. Neuropsychopharmacology 28: 1031–1044.
Holmes A, Picciotto MR (2006). Galanin: a novel therapeutic target for depression, anxiety disorders and drug addiction? CNS Neurol Disord Drug Targets 5: 225–232.
Kaplan LM, Spindel ER, Isselbacher KJ, Chin WW (1988). Tissue-specific expression of the rat galanin gene. Proc Natl Acad Sci USA 85: 1065–1069.
Karatayev O, Barson JR, Chang GQ, Leibowitz SF (2009a). Hypothalamic injection of non-opioid peptides increases gene expression of the opioid enkephalin in hypothalamic and mesolimbic nuclei: possible mechanism underlying their behavioral effects. Peptides 30: 2423–2431.
Karatayev O, Baylan J, Leibowitz SF (2009b). Increased intake of ethanol and dietary fat in galanin overexpressing mice. Alcohol 43: 571–580.
Karatayev O, Baylan J, Weed V, Chang S, Wynick D, Leibowitz SF (2009c). Galanin knockout mice show disturbances in ethanol consumption and expression of hypothalamic peptides that stimulate ethanol intake. Alcohol Clin Exp Res 34: 72–80.
Karlsson RM, Holmes A (2006). Galanin as a modulator of anxiety and depression and a therapeutic target for affective disease. Amino Acids 31: 231–239.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM et al (2002). The human genome browser at UCSC. Genome Res 12: 996–1006.
Kuteeva E, Hokfelt T, Wardi T, Ogren SO (2008a). Galanin, galanin receptor subtypes and depression-like behaviour. Cell Mol Life Sci 65: 1854–1863.
Kuteeva E, Wardi T, Hokfelt T, Ogren SO (2007). Galanin enhances and a galanin antagonist attenuates depression-like behaviour in the rat. Eur Neuropsychopharmacol 17: 64–69.
Kuteeva E, Wardi T, Lundstrom L, Sollenberg U, Langel U, Hokfelt T et al (2008b). Differential role of galanin receptors in the regulation of depression-like behavior and monoamine/stress-related genes at the cell body level. Neuropsychopharmacology 33: 2573–2585.
Leibowitz SF (2005). Regulation and effects of hypothalamic galanin: relation to dietary fat, alcohol ingestion, circulating lipids and energy homeostasis. Neuropeptides 39: 327–332.
Leibowitz SF (2007). Overconsumption of dietary fat and alcohol: mechanisms involving lipids and hypothalamic peptides. Physiol Behav 91: 513–521.
Leibowitz SF, Avena NM, Chang GQ, Karatayev O, Chau DT, Hoebel BG (2003). Ethanol intake increases galanin mRNA in the hypothalamus and withdrawal decreases it. Physiol Behav 79: 103–111.
Leibowitz SF, Kim T (1992). Impact of a galanin antagonist on exogenous galanin and natural patterns of fat ingestion. Brain Res 599: 148–152.
Lu X, Sharkey L, Bartfai T (2007). The brain galanin receptors: targets for novel antidepressant drugs. CNS Neurol Disord Drug Targets 6: 183–192.
Mackenzie A, Miller KA, Collinson JM (2004a). Is there a functional link between gene interdigitation and multi-species conservation of synteny blocks? Bioessays 26: 1217–1224.
MacKenzie A, Purdie L, Davidson D, Collinson M, Hill RE (1997). Two enhancer domains control early aspects of the complex expression pattern of Msx1. Mech Dev 62: 29–40.
MacKenzie A, Quinn JP (2004b). Post-genomic approaches to exploring neuropeptide gene mis-expression in disease. Neuropeptides 38: 1–15.
Madaan V, Wilson DR (2009). Neuropeptides: relevance in treatment of depression and anxiety disorders. Drug News Perspect 22: 319–324.
Mercer JG, Lawrence CB, Atkinson T (1996). Regulation of galanin gene expression in the hypothalamic paraventricular nucleus of the obese Zucker rat by manipulation of dietary macronutrients. Brain Res Mol Brain Res 43: 202–208.
Miller KA, Barrow J, Collinson JM, Davidson S, Lear M, Hill RE et al (2007). A highly conserved Wnt-dependent TCF4 binding site within the proximal enhancer of the anti-myogenic Msx1 gene supports expression within Pax3-expressing limb bud muscle precursor cells. Dev Biol 311: 665–678.
Miller KA, Davidson S, Liaros A, Barrow J, Lear M, Heine D et al (2008). Prediction and characterisation of a highly conserved, remote and cAMP responsive enhancer that regulates Msx1 gene expression in cardiac neural crest and outflow tract. Dev Biol 317: 686–694.
Miller MA, Kolb PE, Raskind MA (1993a). Extra-hypothalamic vasopressin neurons coexpress galanin messenger RNA as shown by double in situ hybridization histochemistry. J Comp Neurol 329: 378–384.
Miller MA, Kolb PE, Raskind MA (1993b). Testosterone regulates galanin gene expression in the bed nucleus of the stria terminalis. Brain Res 611: 338–341.
Mongin E, Dewar K, Blanchette M (2009). Long-range regulation is a major driving force in maintaining genome integrity. BMC Evol Biol 9: 203.
Nagy AG, Vinterstein M, K, Behringer R (2003). Manipulating the Mouse Embryo 3rd edn Cold Spring Harbor Laboratory Press: Cold Spring Harbor.
Naito S, Shimizu S, Matsuu M, Nakashima M, Nakayama T, Yamashita S et al (2002). Ets-1 upregulates matrix metalloproteinase-1 expression through extracellular matrix adhesion in vascular endothelial cells. Biochem Biophys Res Commun 291: 130–138.
Navratilova P, Becker TS (2009a). Genomic regulatory blocks in vertebrates and implications in human disease. Brief Funct Genomic Proteomic 8: 333–342.
Navratilova P, Fredman D, Hawkins TA, Turner K, Lenhard B, Becker TS (2009b). Systematic human/Zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev Biol 327: 526–540.
Ovcharenko I, Nobrega MA, Loots GG, Stubbs L (2004). ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 32 (Web Server issue): W280–W286.
Paschos KA, Veletza S, Chatzaki E (2009). Neuropeptide and sigma receptors as novel therapeutic targets for the pharmacotherapy of depression. CNS Drugs 23: 755–772.
Paxinos KBJFaG (1997). The Mouse Brain in Stereotaxic Coordinates. Academic Press: San Diego, CA.
Picciotto MR, Brabant C, Einstein EB, Kamens HM, Neugebauer NM (2010). Effects of galanin on monoaminergic systems and HPA axis: potential mechanisms underlying the effects of galanin on addiction- and stress-related behaviors. Brain Res 1314: 206–218.
Plaisier CL, Kyttala M, Weissglas-Volkov D, Sinsheimer JS, Huertas-Vazquez A, Riba L et al (2009). Galanin preproprotein is associated with elevated plasma triglycerides. Arterioscler Thromb Vasc Biol 29: 147–152.
Planas B, Kolb PE, Raskind MA, Miller MA (1994a). Activation of galanin pathways across puberty in the male rat: galanin gene expression in the bed nucleus of the stria terminalis and medial amygdala. Neuroscience 63: 851–858.
Planas B, Kolb PE, Raskind MA, Miller MA (1994b). Galanin in the bed nucleus of the stria terminalis and medial amygdala of the rat: lack of sexual dimorphism despite regulation of gene expression across puberty. Endocrinology 134: 1999–2004.
Poritsanos NJ, Mizuno TM, Lautatzis ME, Vrontakis M (2009). Chronic increase of circulating galanin levels induces obesity and marked alterations in lipid metabolism similar to metabolic syndrome. Int J Obes (Lond) 33: 1381–1389.
Rada P, Mark GP, Hoebel BG (1998). Galanin in the hypothalamus raises dopamine and lowers acetylcholine release in the nucleus accumbens: a possible mechanism for hypothalamic initiation of feeding behavior. Brain Res 798: 1–6.
Rokaeus A, Waschek JA (1998). Tissue-specific enhancement and restriction of galanin gene expression in transgenic mice by 5′ flanking sequences. Brain Res Mol Brain Res 60: 150–159.
Rotzinger S, Lovejoy DA, Tan LA (2009). Behavioral effects of neuropeptides in rodent models of depression and anxiety. Peptides 31: 736–756.
Ruano G, Windemuth A, Kocherla M, Holford T, Fernandez ML, Forsythe CE et al (2006). Physiogenomic analysis of weight loss induced by dietary carbohydrate restriction. Nutr Metab (Lond) 3: 20.
Schauble N, Reichwald K, Grassl W, Bechstein H, Muller HC, Scherag A et al (2005). Human galanin (GAL) and galanin 1 receptor (GALR1) variations are not involved in fat intake and early onset obesity. J Nutr 135: 1387–1392.
Scott IG, Akerman KE, Heikkila JE, Kaila K, Andersson LC (1986). Development of a neural phenotype in differentiating ganglion cell-derived human neuroblastoma cells. J Cell Physiol 128: 285–292.
Sergeyev V, Fetissov S, Mathe AA, Jimenez PA, Bartfai T, Mortas P et al (2005). Neuropeptide expression in rats exposed to chronic mild stresses. Psychopharmacology (Berl) 178: 115–124.
Shanley L, Davidson S, Lear M, Thotakura AK, McEwan IJ, Ross RA et al (2010). Long-range regulatory synergy is required to allow control of the TAC1 locus by MEK/ERK signalling in sensory neurones. Neurosignals 18: 173–85.
Shanley L, Lear M, Davidson S, Ross R, Mackenzie A (2011). Evidence for regulatory diversity and auto-regulation at the TAC1 locus in sensory neurones. J Neuroinflammation 8: 10.
Singleton AB, Hardy J, Traynor BJ, Houlden H (2010). Towards a complete resolution of the genetic architecture of disease. Trends Genet 26: 438–442.
Unschuld PG, Ising M, Erhardt A, Lucae S, Kohli M, Kloiber S et al (2008). Polymorphisms in the galanin gene are associated with symptom-severity in female patients suffering from panic disorder. J Affect Disord 105: 177–184.
Unschuld PG, Ising M, Roeske D, Erhardt A, Specht M, Kloiber S et al (2010). Gender-specific association of galanin polymorphisms with HPA-axis dysregulation, symptom severity, and antidepressant treatment response. Neuropsychopharmacology 35: 1583–1592.
Vetter M, Blumenthal SG, Lindemann RK, Manns J, Wesselborg S, Thomssen C et al (2005). Ets1 is an effector of protein kinase Calpha in cancer cells. Oncogene 24: 650–661.
Wray GA (2007). The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216.
Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR et al (2010). Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry.
Wynick D, Bacon A (2002). Targeted disruption of galanin: new insights from knock-out studies. Neuropeptides 36: 132–144.
Yee SP, Rigby PW (1993). The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev 7: 1277–1289.
Acknowledgements
This work was funded by the Tenovus Trust, Scotland, the BBSRC (BB/D004659/1), the Wellcome Trust (080980/Z/06/Z) and the Medical Research Council (G0701003). SD was funded by a BBSRC strategic studentship (BBS/S/2005/12001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Davidson, S., Lear, M., Shanley, L. et al. Differential Activity by Polymorphic Variants of a Remote Enhancer that Supports Galanin Expression in the Hypothalamus and Amygdala: Implications for Obesity, Depression and Alcoholism. Neuropsychopharmacol 36, 2211–2221 (2011). https://doi.org/10.1038/npp.2011.93
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2011.93
Keywords
This article is cited by
-
An ancient polymorphic regulatory region within the BDNF gene associated with obesity modulates anxiety-like behaviour in mice and humans
Molecular Psychiatry (2024)
-
The anxiety and ethanol intake controlling GAL5.1 enhancer is epigenetically modulated by, and controls preference for, high-fat diet
Cellular and Molecular Life Sciences (2021)
-
Neuropeptide signalling in the central nucleus of the amygdala
Cell and Tissue Research (2019)
-
Oprm1 A112G, a single nucleotide polymorphism, alters expression of stress-responsive genes in multiple brain regions in male and female mice
Psychopharmacology (2018)
-
Analysis of the effects of depression associated polymorphisms on the activity of the BICC1 promoter in amygdala neurones
The Pharmacogenomics Journal (2016)