Abstract
A recently discovered gene complex, G72/G30 (hereafter G72, but now termed DAOA), was found to be associated with schizophrenia and with bipolar disorder, possibly because of an indirect effect on NMDA neurotransmission. In principle, if G72 increases risk for psychosis by this mechanism, it might impact with greater penetrance those cortically based cognitive and neurophysiological functions associated with NMDA signaling. We performed two independent family-based association studies (one sample contained more than 200 families and the other more than 65) of multiple SNPs in the G72 region and of multiple SNPs in the gene for D-amino acid oxidase (DAAO), which may be modulated by G72. We examined the relationship between select cognitive measures in attention, working memory, and episodic memory and a restricted set of G72 SNPs in over 600 normal controls, schizophrenic patients, and their nonpsychotic siblings using mixed model ANOVAs. We also determined genotype effects on neurophysiology measures in normal controls using the fMRI BOLD response obtained during activation procedures involving either episodic memory or working memory. There were no significant single G72 SNP associations and clinical diagnosis in either sample, though one approached significance (p=0.06). Diagnosis by genotype interaction effects for G72 SNP 10 were significant for cognitive variables assessing working memory and attention (p=0.05), and at the trend level for episodic memory, such that in the schizophrenia group an exaggerated allele load effect in the predicted directions was observed. In the fMRI paradigms, a strong effect of G72 SNP 10 genotype was observed on BOLD activation in the hippocampus during the episodic memory paradigm. Tests of association with DAAO were consistently nonsignificant. We present evidence that SNP variations in the G72 gene region increase risk of cognitive impairment in schizophrenia. SNP variations were not strongly associated with clinical diagnosis in family-based analyses.
Similar content being viewed by others
INTRODUCTION
Over the past several years, the search for genetic causes of schizophrenia has yielded several linkage regions from which candidate genes have emerged. One such candidate, identified as G72 (and now termed DAOA) was recently found in a ‘gene desert’ after construction of a dense marker map in a five megabase region on chromosome 13q, which had been previously identified as a site of suggestive or significant linkage (Chumakov et al, 2002). Following annotation and in vitro translation, the gene was shown to code a 150+ amino-acid protein. G72 appears to be related to a larger gene on the opposite DNA strand, which has been called G30. Multiple SNPs in this region were found to be associated with schizophrenia in two large samples, one of French-Canadian descent, the other of Russian descent. Biochemical experiments revealed that the G72 protein activated a second protein, D-amino acid oxidase (DAAO), involved in the metabolism of D-serine, an agonist at the glycine modulation site of the NMDA receptor (summarized in Harrison and Owen, 2003). Several SNPs in the region of the DAAO gene were also found to be significantly associated with schizophrenia in the French-Canadian sample. G72's protein was found to be expressed in brain at low levels and to have numerous splice variants. A second study, from an independent group (Hattori et al, 2003), using both individual SNP markers and multimarker haplotype analyses, found that variation in G72 was significantly associated with bipolar disorder in two series of pedigrees. A case control study in which no population stratification was discerned also found an association of G72 to bipolar disorder, although not in the SNPs previously studied (Chen et al, 2004). A recent large case control study found evidence of association for select SNPs and a haplotype with both schizophrenia and bipolar disorder (Schumacher et al, 2004). Last, G72 SNPs were associated with psychotic illnesses in children (Addington et al, 2004). However, positive SNPs were not always consistent among the studies. Functional variations in the gene have yet to be identified.
The association of G72 with psychosis and the potential relationship between G72 and NMDA function are consistent with several lines of evidence suggesting that cortical glutamatergic neurotransmission may be compromised in schizophrenia (for reviews see Goff and Coyle, 2001; Tsai and Coyle, 2002; Krystal et al, 2002): (1) there is neuropathologic evidence of glutamate receptor abnormalities in prefrontal and temporal cortices that has been described in studies of post-mortem tissue of patients with schizophrenia (Deakin et al, 1989; Tsai et al, 1995; Meador-Woodruff and Healy, 2000); (2) drugs that reduce glutamatergic transmission at NMDA receptors in normal human subjects, such as ketamine, produce a range of cognitive impairments, including those in attention/working memory, word generation, and episodic memory that are somewhat similar to those found in patients with schizophrenia, as well as a range of symptoms often found in schizophrenia (Krystal et al, 2000; Krystal et al, 1998; Malhotra et al, 1996); and (3) animal models that involve knockdowns of NMDA receptor subunit genes (Gainetdinov et al, 2001) or chronic administration of NMDA antagonists (Jentsch and Roth, 1999) have produced animals whose behavior and brain function are in some respects ‘schizophrenia-like.’ However, it remains unclear if differences between long-term and short-term modulation of the glycine site are present (Lin et al, 1998) or if effects of G72 are age-specific.
In this study, we have examined the relationship of SNP variations in G72 and in DAAO and risk for schizophrenia in two family association data sets of American ethnicity and also explored the potential relationships between variation in these genes and aspects of human cortical function related to glutamate activity. We reasoned that if G72 increases risk for psychosis by virtue of its putative role as an indirect modulator of NMDA neurotransmission, it should also impact on cortically based cognitive functions associated with NMDA signaling, which also have been associated with psychosis.
METHODS
General
Subjects
Subjects were recruited as part of the Clinical Brain Disorders Branch NIMH ‘Sibling Study’ and included a group of healthy controls, a group of patients with broadly defined schizophrenia that included diagnoses of schizophrenia, schizoaffective disorder, delusional disorder, and schizophrenic spectrum personality disorders (schizotypal, schizoid, and paranoid, that is, cluster A) and hereafter called the schizophrenia affected group, and a group of siblings of these index cases who did not carry such diagnoses. All diagnoses were made following SCID interviews. To reduce the possibility of population stratification confounds, all analyses were performed in subjects from a common ethnic group, namely Americans of European extraction, that is, Caucasian (according to self reports). Prior reports of association to G72 also have been in Caucasian samples. Owing to the prior association of G72 with bipolar disorder, we also constructed an additional diagnostic group of subjects consisting of bipolar patients, manic type added to the schizophrenia group (called the psychosis group). Subjects were screened for systemic and neurological conditions that might impact brain function (eg head injury, dyslexia, attention deficit disorders, current alcohol, or substance abuse, type 2 diabetes). Ascertainment procedures are reviewed in Egan et al, 2001a, 2001b.
A replication sample for family-based association analysis was obtained from the NIMH Genetics Initiative (NIMHGI) data set (Cloninger et al, 1998). Caucasian families were selected that included at least two siblings with a diagnosis of schizophrenia or schizoaffective disorder, depressed and at least one parent. Details of ascertainment, diagnostic procedures, and DNA collection have been described (Cloninger et al, 1998).
Genotyping
G72 and DAAO genotypes were determined using the Taqman exonuclease assay. All subjects were genotyped in the G72 region of chromosome 13q and the DAAO region of chromosome 12q. Eleven G72 SNPs were examined. All had been provided by Chumakov et al (2002): nine were included in published work, whereas two were communicated (Ilya Chumakov, personal communication). One SNP typed by Chumakov et al could not be reliably genotyped due to technical problems (M 12) and was omitted. Four SNPs of the set that we analyzed were also examined by Hattori et al, 2003. Nomenclature is listed in Table 1. Five DAAO SNPS were typed (MDAAO 3–7, using the nomenclature of Chumakov et al). DNA was available from schizophrenia patients, their siblings, and biological parents, and healthy controls. Nucleotide alleles mentioned herein refer to the coding strand.
All SNPs were in Hardy–Weinberger equilibrium (HWE) in the patients and in controls. TRANSMIT was used to identify families with non-Mendelian inheritance. All individuals in those marker-family combinations were set to genotype unknown.
The completion rate for genotyping was near 90%. The rate of genotyping error for the reported SNPs (as detected during data cleaning as opposed to the general lab error) is as follows. For the 11s, the number of non-Mendelizations that TRANSMIT reported ranged from 4 to 13, an average of 6.5 per marker. Almost invariably, we have found that this represents one genotyping error per family, so given the 935 family members being genotyped, we estimate a range of 4/935 (0.4%) to 13/935 (1.4%). Note that these are the errors detected—the actual rate is of course somewhat higher. Overall, we have found that for markers performing well (ie, good clusters) in the Taqman assay, the Taqman genotype discordance from the genotype determined from double-stranded sequencing is <1%. Reproducibility of Taqman genotypes is in general >99.5%.
Regarding allele frequencies, Hattori et al (2003) did not provide allele frequencies. The minor allele frequencies for the Canadian sample in Chumakov et al (2002) were always within 4% of our frequencies, with the exception of M-22 (our marker 8, rs778 293), where the frequency in the Canadian sample was 31% in cases and 40% in controls, whereas in our sample it was 43% overall in families.
Regarding haplotypes used in the association studies, we analyzed consecutive, overlapping three marker haplotype blocks (ie, sliding window). There were thus nine blocks.
The databases used herein have also been used to examine associations with the following candidate genes: COMT, BDNF, GRM3, and DISC1.
Study 1
G72 and DAAO SNP transmission disequilibrium analyses
Subjects: Across all G72 and DAAO SNPs, the number of families with transmissions to ill offspring in the CBDB data set ranged from 199 to 217 and the number of ill offspring ranged from 211 to 230. For SNP 7, the SNP with the largest number of families with transmissions, 106 trios were fully typed, 46 ‘trios’ had data from one parent and at least one sibling, and 65 ‘trios’ had sibling data but no parental data. All other SNPs had similar proportions of complete and incomplete family data. Of the families (all Caucasian), 67 were included from the NIMHGI sample. Those SNPs tested in the CBDB dat set were also tested in the NIMHGI sample.
Statistics: TRANSMIT, a nonparametric method of determining association based on transmission of alleles within families (Clayton, 2001), was used to assess if individual SNPs or successive three marker haplotypes were over or undertransmitted from parents to schizophrenia offspring. If parental data were incomplete, the missing genotypes were reconstructed from the available family data (including second degree relatives in some NIMHGI cases) where possible. The haplotype analyses were restricted to common haplotypes (>3% frequency). Additionally, pairwise LD between SNPs was computed.
Results: One individual G72 SNP demonstrated near significantly distorted transmission from parents to schizophrenia offspring by TRANSMIT analyses: SNP 1 (p=0.06); the G nucleotide (coding strand) was overtransmitted. These results are detailed in Table 1. When bipolar patients were included (in the ‘psychosis’ group), results were negative for all SNPs. Details of the analyses are available upon request.
One three marker haploptype, consisting of alleles C, T, and T at SNPs 5/6/7 reached borderline significance (X2=3.71, p=0.09 maximum; X2=9.17, p=0.02 global) in the broad diagnostic group. Approximately, six overtransmissions were observed (26.6 observed vs 21.1expected). All other three marker haplotypes were negative (p-values>0.10).
No individual SNP or three marker haplotype demonstrated significant overtransmission from parents to schizophrenia offspring in the NIMHGI study by TRANSMIT. These results are available upon request.
No individual DAAO SNP or multimarker haplotype demonstrated significantly distorted transmission for broadly defined schizophrenia offspring. For MDAAO SNP 3 X2=0.11, p=0.73, for MDAAO 4 X2=0.39, p=0.53, for MDAAO 5 X2=0.04, p=0.85; MDAAO 6 X2=0.001, p=0.97; and for MDAAO 7 X2=0.09, p=0.77.
Pairwise LD among G72 SNPs indicated the presence of two blocks of highly intercorrelated SNPS: 3–7 and 8–10, as displayed in Table 2.
Study 2
Association of G72 and DAAO genotypes to cognitive function
To reduce the confound of multiple testing, we focused on two SNPs from the Chumakov study that were significant in both their samples (at p<0.02) and were also positive in the Schumacher et al (2004) schizophrenia cases (SNPs 9 and 10). In each, the T nucleotide was associated with schizophrenia in their study and we posited that the Ts would be cognitively disadvantageous in our sample. We also examined a third SNP (7) that was not significant in the Chumakov et al (2002) samples; we considered this a ‘negative’ control. Last, we examined a SNP (5) that in the Hattori et al (2003) study was significant in both of their pedigree data sets. Similarly, we examined those three DAAO SNPs from the Chumakov et al study that showed a positive association to schizophrenia (DAAO 5–7).
Subjects: Demographic information for the 600+ subjects is listed in Table 3 by group. Of the affected subjects all but 17 were receiving neuroleptic medication. For cognitive phenotypes, the Ns are as follows: for the N Back 123 controls, 124 siblings, 81 probands, for all other tests 150+ controls, 245+ sibs, and 180+ probands.
Cognitive tests: The cognitive tests utilized in this study were selected for analyses because they: (1) had been implicated as schizophrenia-related intermediate phenotypes in earlier family studies based on increased relative risk measures or significant parametric statistics (N Back, letter fluency, memory for stories, paired associates, IQ, Trails B) (Egan et al, 2001a, 2001b; Goldberg et al, in press); (2) were known to be sensitive to the effects of glutamate manipulations (WCST, letter fluency, CPT) (Malhotra et al, 1996; Krystal et al, 2002; Krystal et al, 1994), or (3) were important predictors of clinical outcome (WCST, IQ, memory for stories and paired associates, CPT) (Goldberg and Green, 2002; Goldberg et al, 1990). They were administered using standardized procedures and were part of a larger battery of tests. The tests are as follows and are described in detail elsewhere (Egan et al, 2001a, 2001b; Goldberg et al, 2003).
N Back Working Memory Test (One Back): measures working memory subprocesses involving attention and ‘updating’ of information. Performance was expressed as percent correct.
Wisconsin Card Sorting Test: executive function during a card sorting task. The number of perseverative errors was considered the critical dependent measure.
Continuous Performance Test (CPT) Distractibility Version: attention and vigilance to numeric stimuli in the presence of interfering stimuli. Performance was expressed by d’.
Memory for Stories: verbal episodic memory. The number of story elements recalled was the index of performance.
Verbal Paired Associate Learning (Hard Pairs; from the Wechsler Memory Scale-Revised): episodic learning of unrelated word pairs over multiple trials. The number of words recalled over three trials was the index of performance.
Verbal Fluency (Letters): lexical/semantic memory processing during timed word generation. The number of words produced over three 1 min periods was the measure of performance.
Trail Making (Trails B): oculomotor scanning and attention. Time in seconds to complete the test was the dependent measure.
WAIS-R IQ: ‘intelligence’ The short form used here was comprised of the subtests of Picture Completion, Similarities, Digit Symbol, and Arithmetic (Kaufman, 1990).
Statistics: Parametric analyses utilized a rigorous mixed-effects ANOVA in which a family was treated as a random effect in order to account for nonindependence of unaffected and affected pairs (Littell et al, 1996). Main effects for diagnostic group (healthy controls, siblings, and schizophrenia index cases) and genotype were examined, as were genotype × diagnosis interactions.
Association of G72 and DAAO genotypes to cognitive function
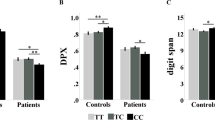
SNP 10 yielded significant main effects of genotype or genotype × diagnosis interactions on the following cognitive variables: CPT, One Back, and Verbal Paired Associates (see Table 4). Effects are illustrated in Figure 1. Univariate contrasts within the schizophrenia group indicated significant differences between T/T and A/A genotype (by t-test, all p's<0.05) for these three cognitive measures. Significant effects of SNP 10 on other cognitive variables (IQ, WCST, verbal fluency, memory for stories, Trails b) were not observed. Regarding other possible disease-related associations that might have skewed cognitive findings, no associations between SNP 10 genotype and schizophrenia subtype diagnosis and type of medication (typical and atypical) were observed (p=0.59 and p=0.35 by χ2, respectively).
Performance as a function of diagnosis and G72 SNP 10 genotype in select cognitive tests. (a) Accuracy scores on the One Back test of working memory demonstrating a significant main effect of G72 10 genotype and a significant interaction such that A,A homozygotes performed better than T,T homozygotes notably in the schizophrenia group (Normal TT N=24, Normal AT N=54, Normal AA=19; Sib TT N=36, Sib AT N=41, Sib AA N=22; Schizophrenia TT N=19, Schizophrenia AT N=42, Schizophrenia AA N=13). (b) Accuracy scores on the CPT test of attention demonstrating a significant main effect of G72 10 genotype and significant interactions such that A,A homozygotes performed better than T,T homozygotes notably in the schizophrenia group. (Normal TT N=31, Normal AT N=64, Normal AA=28; Sib TT N=64, Sib AT N=85, Sib AA N=41; Schizophrenia TT N=43, Schizophrenia AT N=82, Schizophrenia AA N=30). (c) Accuracy scores on the Verbal Paired Associates Learning test of episodic memory demonstrating a significant main effect of G72 SNP 10 genotype and a near significant interactions such that A,A homozygotes performed better than T,T homozygotes (Normal TT N=30, Normal AT N=64, Normal AA=28; Sib TT N=58, Sib AT N=77, Sib AA N=38; Schizophrenia TT N=42, Schizophrenia AT N=81, Schizophrenia AA N=29). Error bars reflect SEMs.
SNP 9 yielded a significant genotype × diagnosis interaction on the One Back task (F=3.84, p=0.007). Trends (p<0.10) for significant interactions were also observed on Verbal Paired associates and the CPT. Like SNP 10, schizophrenia individuals with the T,T genotype performed worse than those with the A,A genotype in all three measures (CPT, One Back, Verbal Paired Associate Learning).
Correlations between CPT and One Back were significant in all groups; correlations between these variables and verbal paired associate learning were low and nonsignificant, suggesting that G72 10 influences multiple cognitive domains (ie, working memory/attention and episodic memory).
No significant genotype effects or genotype × diagnosis interactions were observed in any cognitive variable for the negative control SNP, that is, 7, or for the SNP chosen from the study of Hattori et al (2003). No significant genotype effects or genotype × diagnosis interactions were observed in any cognitive variable for any of the three DAAO SNPS (5–7).
Given the large number of analyses, we used the Benjamini Hochberg procedure for control of false discovery rate as implemented by SAS PROC Multtest. We applied FDR to the cognitive data set for SNPs 10 and 7, which were not in pairwise LD, and included all cognitive measures. The One Back remained significant; (p=0.04) for SNP 10; CPT and verbal paired associate learning demonstrated trends (p<0.08).
Study 3
FMRI studies of G72 SNP 10
General: The fMRI BOLD response is a more powerful method of capturing genotypic effects than cognition alone, both because of its use of time series data and its use of a dependent variable that is nearer to the neurobiology of the gene; that is, penetrance should be greater at the level of brain information processing than at the behavioral level that is cognition (Gabrieli and Preston, 2003; Hariri and Weinberger, 2003). Therefore, we also looked for association with phenotypes derived from functional neuroimaging in normal subjects (we did not have an adequate sample of genotyped schizophrenic individuals who had useable fMRI data). In the functional neuroimaging studies which follow, we focused our experiments on G72 SNP 10, in order to provide convergent evidence for its effects on critical aspects of cognition involving attention/working memory and episodic memory. We reasoned that genotypic effects on regional neurophysiology in healthy controls might be identified in these paradigms.
Whole brain BOLD fMRI data were collected on a 3 Tesla Signa scanner using a GE-EPI RT pulse sequence (GE, Milwaukee, WI, USA) acquiring 24 interleaved slices (echo time=30 ms, repetition time=2 s, flip angle=90°, field of view=24 cm, matrix=64 × 64, voxel dimensions 3.75 × 3.75 × 6 mm) covering the whole brain. All fMRI data were processed and spatially normalized to a common stereotaxic space (Montreal Neurologic Institute template) and analyzed using a random effects model within SPM99 software (Brett, 2004; Wellcome Department of Cognitive Neurology, 2000).
Study 3a. FMRI study of G72 regulation of hippocampal engagement during episodic memory
Based on the genotypic impact on verbal paired associate learning, we predicted that individuals homozygous for the T allele (which was the ‘disadvantageous’ genotype in the schizophrenia group in terms of cognitive penetrance) would show abnormal temporal cortical function during encoding of to-be-remembered items than individuals with A,T or A,A genotypes. Prior studies using fMRI during declarative memory indicate that memory function covaries with degree of hippocampal activation (Hariri et al, 2003), so it would be predicted that T,T individuals would show lesser hippocampal activation.
Subjects: Seven normal individuals homozygous for the T allele at SNP 10 (ie T,T, the putatively disadvantageous genotype) were compared to seven individuals carrying an A allele (ie A,T and A,A genotypes). IQ was virtually identical between the groups. The two groups did not differ in frequencies of BDNF genotypes or of APOE4 genotypes (two genes previously found to be associated with hippocampal activation during episodic memory (Egan et al, 2003; Burggren et al, 2002)). Neither age, handedness, nor sex ratio differed between the two groups.
Methods We examined medial temporal lobe physiology with fMRI during a cognitive paradigm involving encoding of novel affectively neutral complex indoor and outdoor visual scenes, followed by recognition of these scenes as described in detail elsewhere (Hariri et al, 2003). Previously this task has been shown to elicit activation at both encoding and recognition in parahippocampal and hippocampal regions. We also performed a second similar experiment using affectively toned aversive indoor and outdoor scenes (eg, car crash). All stimuli came from the International Affective Picture System (IAPS) (Lang et al, 1997). Stimuli for both experiments were presented in four interleaved encoding and four interleaved retrieval blocks with a passive rest condition. During encoding blocks, subjects viewed six images and determined whether they were ‘indoor’ or ‘outdoor’ scenes. During subsequent retrieval blocks, subjects again viewed six images, presented serially for 3 s each, and determined if each scene was ‘new’ or ‘old’. In each retrieval block, half the scenes were ‘old’ (ie presented during the encoding blocks) and half were ‘new’ (ie not presented during the encoding blocks). During interleaved rest blocks, subjects fixated on a central crosshair. Prior to the beginning of each block, subjects viewed a brief (2 s) instruction: ‘Indoor or Outdoor?’, ‘New or Old?’, or ‘Rest’. During scanning, all subjects responded by button presses with their right hand. Statistical analyses determined effects within each voxel in an anatomically defined region that included the right and left hippocampi and parahippocampal cortices using the method of Giedd et al (1996). First a t-statistic calculated contrasts of encoding vs rest and retrieval vs rest for each subject. These contrast images were used in a second level random effects model to determine differences in task-specific BOLD responses at the group level and t-tests to determine differences between the two genotype groups in a region of interest (here medial temporal lobe), with small volume correction. Coordinates for significant activation clusters were converted to the standard space of Talaraich and Tournoux (1988) and reported as local maxima.
Results In the experiment using neutral scenes, greater BOLD signal in the A carrier group in comparison to the T,T group was observed in a region in the left parahippocampal gyrus (BA 36/37) during encoding (Talaraich coordinates −26, −48, −6; 4 voxels; Z=2.09, p=0.04 svc). Genotype-based statistical parametric maps that illustrate significantly greater activation in the A carrier group can be seen in Figure 2. No differences in BOLD response between the groups were observed during recognition in the medial temporal lobe region. Behavioral results revealed no significant differences in performance at recognition between the A carrier and T,T groups (error x=2.50±1.19 vs 3.25±2.76, t=0.70, p=0.49).
SPM parametric maps are shown overlaid onto an averaged structural MRI for comparisons between G72 10 genotypes for memory of neutral scenes. Genotype-based parametric comparisons of BOLD fMRI data demonstrating significantly greater left parahippocampal activation during encoding of neutral scenes by the A carrier group vs the T,T group.
In the experiment using aversive scenes, greater BOLD signal in the A carrier group in comparison to the T,T group was observed in locales in the left parahippocampal gyrus (BA 37) and right hippocampus during encoding (Talaraich coordinates −29, −44, −6; 5 voxels; Z=2.32, p=0.03 svc and 26, −26, −7; 4 voxels Z=1.86, p=0.04 svc). During recognition greater BOLD signal was observed in the A carrier group in comparison to the T,T group in regions of the right parahippocampal gyrus (Talaraich coordinates 34, −48, −6, 20 voxels; Z=3.51; p=0.01 svc). Thus, the aversive stimuli resulted in genotype-based differences at both encoding and recognition that were in the predicted directions. Genotype-based statistical parametric maps illustrate significantly greater activation in the A carrier group in Figures 3a and b. Behavioral results favored the A carrier group, but not significantly (error x=1.14±1.67 vs 1.76±1.97, t=0.59, p=0.67). Owing to the small sample size these results require replication.
SPM parametric maps are shown overlaid onto averaged structural MRIs for comparisons between G72 SNP 10 genotypes for aversive scenes (a). Genotype-based parametric comparisons of BOLD fMRI data demonstrating significantly greater left parahippocampal and right hippocampal activation during encoding of aversive scenes by the A carrier group vs the T,T group. (b) Genotype-based parametric comparisons of BOLD fMRI data demonstrating significantly greater right parahippocampal activation during retrieval of aversive scenes by the A carrier group vs the T,T group.
Study 3b. FMRI study of G72 regulation of frontal cortex engagement during working memory
fMRI BOLD response in prefrontal cortex was determined during the N Back working memory task (Callicott et al, 2000). The N Back task was used as a cognitive challenge because it reliably activates a working memory and attentional network that includes prefrontal cortex. Critically, in groups known to have prefrontal cortical pathophysiology (eg schizophrenia, Parkinson's disease), an inefficient response characterized by greater BOLD activation during this version of the Two Back test (vs 0 Back) has been found (Callicott et al, 2000; Mattay et al, 2002). Thus, we predicted that the individuals with the disadvantageous homozygous T,T genotype would show a less efficient neurophysiological response to working memory demands and vigilance in prefrontal cortex than individuals with A,T or A,A genotypes as based on prior studies.
Subjects: Ten normal individuals with the putatively disadvantageous T,T homozygous genotype were compared to 20 normal individuals carrying an A allele at SNP 10 (ie A,T and A,A genotypes). Importantly, subjects were matched on N Back accuracy to remove variance due to performance, per se. (The accuracy of both groups in the Two Back task was approximately 83%.) Differences in BOLD response thus presumably reflected variation in neural processing, rather than cognitive ability, strategy, or test taking style. The groups were also equivalent in handedness scores, age, and sex to reduce confounding factors.
Methods: Subjects who performed the N Back working memory task using a block design that alternated between a Two Back and a control condition (the Zero Back). During the Two Back working memory condition, subjects were required to recall a number (digits 1, 2, 3, or 4) seen two stimuli previously. During the Zero Back, subjects identified the number currently on the screen. Visual stimuli were presented via a back-projection screen and performance (accuracy and reaction time) recorded via the use of a button box held in the right hand. Single subject contrast maps were created by comparing Two Back working memory and Zero Back control tasks blocks for voxels showing significant change in BOLD signal using a one sample t-test (p<0.01, corrected). All individual maps showed activation of the working memory network. A mask of activated regions was created from the summation of individual subjects’ activation maps. Next, a group comparison of genotype was then performed as a second level (random effects) one way ANOVA within SPM 99 (with p<0.01, uncorrected; cluster size k>5). Coordinates for significant activation clusters were converted to the standard space of Talairach and Tournoux and reported as local maxima.
Results: Greater BOLD activation in the T,T group was found in right superior prefrontal cortex (middle frontal gyrus) BA 6 (30, 21, 54, 5 voxels, Z=2.70), a right-sided prefrontal subgyral area (19, 29, −1, 9 voxels, Z=2.72) and cerebellar regions (−8, −56, −33; 4, −39, −33; −22, −49, 28). The A carrier group, however, demonstrated greater BOLD activation in left frontal cortex (precentral gyrus) BA 4/6 (−49, 2, 50, 5 voxels, Z=2.88), right caudate (11,1,11, 9 voxels, Z=3.12) and right insula (41, 11, 5, 8 voxels, Z=2.65). The differences in the genotype groups found in the second level analysis were outside predicted networks.
DISCUSSION
We performed a family association study of schizophrenia using SNPs in the G72/G30 gene region in two independent samples of American-Caucasian families. We did not find significant association of individual markers or of multimarker haplotypes to schizophrenia in either group of families; however our study samples of trios were not large. We did find evidence, however, of a genotype by diagnosis interaction with cognitive intermediate phenotypes that are associated with schizophrenia. Specifically, increasing load of an allele associated with schizophrenia in two case control samples reported by Chumakov et al (2002), that is, at SNP 10—amplified the burden of cognitive impairment in a linear fashion in patients with schizophrenia. Similar results were found for the other positive SNP (9) from Chumakov et al, but not for a ‘negative’ SNP from their study. Consistent with our conjecture that there would be greater penetrance of gene effects at the level of brain function than at clinical phenomena, we also found that the same allele of SNP 10 was associated with quantitative variance in brain physiology in normal subjects in the medial temporal lobe and that the directionality of the association was consistent with the information processing deficits associated with schizophrenia. Thus, while there was little penetrance of G72 SNPs at the clinical diagnostic level in our cohort, penetrance at the level of cortical function was observed in the schizophrenia group using cognitive performance and in the normal group with fMRI.
One way to interpret these data, at least in the patients, is to propose an interaction or epistasis model. In general, SNP 10 genotype effects on cognition were greatest in schizophrenic index cases such that individuals with the T,T genotype had the poorest performance and cases with A,A genotype the best performance. The findings were less consistent in siblings and in healthy controls, although we note that a main effect of genotype was nevertheless present suggesting at the very least that these groups did not markedly attenuate the primary finding. It thus appears that the G72 polymorphism at SNP 10 (or a variation in linkage disequilibrium with this marker), in the context of the genetic architecture of schizophrenia, has relatively large cognitive effects. However, because nearly all schizophrenic patients were receiving neuroleptic medications, it cannot be ruled out that we were observing a ‘genotype × neuroleptic’ interaction.
Nevertheless, the effects were observed in those very cognitive processes that appear to be both heritable and modal in schizophrenia (Egan et al, 2001a; Weickert et al, 2000) and that the effect was amplified in schizophrenia, suggests that G72 genotype may be an epistatic risk factor that increases the likelihood of schizophrenia type cognitive impairment in individuals with other risk factors. In other words, the genetic background against which variation in this gene occurs may determine the magnitude of its impact. We also have preliminary data indicating that G72 interacts with a functional COMT SNP to amplify cognitive impairments (Goldberg, Egan, Straub, Weinberger, unpublished). Presumably, with the accumulation of disadvantageous alleles, it becomes more difficult to compensate for the deleterious effects of any one specific allele. Alternatively the SNPs used in this study were in monitoring relations to another perhaps relatively rare causative mutation that was enriched in the schizophrenic sample.
In addition to potential epistatic effects in patients, subtle yet discernible effects on hippocampal neurophysiology were observed in healthy controls using fMRI during memory encoding tasks. This further supports the assumption that susceptibility risk genes for schizophrenia show greater penetrance at the level of brain information processing than at the level of behavior. Perhaps it is in some sense ‘easier’ for an individual to compensate for the presence of a disadvantageous allele at the behavioral level, using alternative cognitive strategies, response monitoring, instructional set, or effort than at the physiological level.
In one fMRI study (involving the N Back), we considered greater activation to reflect inefficiency whereas in the medial temporal lobe we considered greater activation to be associated with better functioning. Using the N Back (in which a response is made to every stimulus) and in which groups have been matched on performance, we have noted ‘inefficiency’ in studies involving individuals with compromised frontal lobe function, including patients with schizophrenia, Parkinson's disease patients, normal elderly, and normal subjects with the COMT val/val genotype. In contrast, the episodic memory task involving visual scenes has elicited reduced activation in both normal subjects carrying the BDNF met allele and schizophrenic individuals who have impaired memory function.
Our results, taken in toto, implicate significant effects of G72 in prefrontal and medial temporal neural systems that are reliably engaged in working memory and episodic memory, respectively. As G72 is thought to be an indirect modulator of NMDA receptor function (Harrison and Owen, 2003) it would not be unexpected for variation in the gene to have effects in these neural systems, and the cognitive domains that engage them. However, the neurobiological data on G72 is scant and our perspective on G72's role in glutamate neurotransmission is a working hypothesis. Certainly, G72 does not have the same magnitude of impact on cognitive function produced by direct pharmacological manipulation of the NMDA receptor by ketamine blockade.
Given the significant findings of G72 effects on N Back and CPT performance, the gene likely has effects on cognitive processes common to both tasks, for example, attentional processes occurring within working memory that involve target detection or stimulus encoding and response (Elvevaag et al, 2000). It is also interesting to note that this type of demand for cognitive precision at the representational level has been found to be sensitive to glutamatergic manipulation at NMDA receptors (Schroeder et al, 2001; Doniger et al, 2001).
The episodic memory task that was sensitive to G72 regulation is reliably impaired in a variety of memory disorders, especially when difficult paired associates must be remembered. Although the association of G72 and verbal paired associate learning was at the trend level, it is consistent with other studies examining the role of NMDA function and memory and our own fMRI study (3a). The paradigm requires binding of arbitrary elements into an episode with a distinct spatiotemporal context; this is a critical feature of medial temporal lobe engagement in cognitive processing. In fact, glutamatergic transmission at NMDA receptors is thought to be a prerequisite for Ca++ influx, which initiates an intracellular signaling cascade that is necessary for LTP and putatively the formation of memories in which disparate sensory elements are bound into a single episode (Pittenger and Kandel, 2003).
The most consistent evidence for G72's association with cognitive impairment was in two SNPs near the 3′ end of the gene (9 and especially 10). Other SNPs that we examined yielded neither significant differences in genotype, nor significant genotype by diagnostic group interactions. SNP 10 was one of only two SNPs (the other being 9) found by Chumakov to show significant allelic associations in both their Russian and Canadian samples. In SNP 10 (and in SNP 9) the T nucleotide, which was associated with schizophrenia by Chumakov et al and Schumacher et al (2004), was the variant associated with worse cognitive performance in the schizophrenic group in this study. We have no evidence that this allele impacts on gene function; it is possible that it is in a monitoring relationship with an as yet to be determined causative mutation or other variant. Last, it is interesting to note that the disadvantageous T allele of SNP 10 (marginally more frequent that the A allele in all groups) is the common allele at this locus. This is consistent with much recent work that suggests that common alleles can be associated with diseases of complex genetic etiology (Egan et al, 2001b; Cox, 2002; Lohmueller et al, 2003).
Although we might claim to have found weak evidence that G72 is a susceptibility gene for schizophrenia as a diagnostic entity in this sample, we suggest that our results are negative, that is, not significant. None of 11 TRANSMIT analyses of individual SNPs was of significance (although one SNP was at the trend level). A single haplotype was significant at a trend level. However, these were removed from the positive cognitive SNPs located at the 3′ end of the gene. We also did not find significant association of individual SNPs to schizophrenia using a more restricted diagnostic scheme (ie as Chumakov et al (2002) apparently utilized) or with a more global psychosis scheme. We do not have an explanation for our negative clinical findings in comparison to those of Chumakov et al, although we do not think that our cohort is unrepresentative of the population of schizophrenic individuals, either in terms of symptoms or cognitive impairments. Clearly, however, our family-based association study has limited power. Genetic power analyses (using Statgen (Purcell, 2003)) indicated that for SNP 1, which approached significance by TDT, a sample of 331 trios would be expected to yield significance at 0.80 power, based on observed and expected transmissions and allele frequency in our sample. When we conducted a power analysis for the individual SNP that demonstrated the greatest association with cognitive variables (SNP 10) the difference in expected and observed was very small (ie 230 vs 231) and so would literally necessitate a sample of trios in the thousands to obtain significance.
There has been recent evidence for a role for G72 in the etiology of another psychotic neuropsychiatric disorder, namely bipolar disorder, but the impact of SNPs on cognition in bipolar disorder is unknown. Moreover, the SNPs that showed significant association in the bipolar samples were negative in our sample and did not relate to cognition.
We also found no evidence for genetic association of DAAO SNPs either with schizophrenia or with cognitive phenotypes. Relatively little molecular and genetic work has been conducted on this gene and its neurobiological effects are not well understood.
Statistical issues associated with multiple comparisons also need to be addressed. We chose a variety of tasks from multiple cognitive domains because we were unsure which measures would be most sensitive to the cognitive processes that we putatively assigned to G72 genotypes, given the lack of prior work in this area. This being said, several of the tasks may be intercorrelated, in essence, reducing the number of independent comparisons (eg N back and CPT). Similarly, each SNP is not independent from every other SNP. Some are in linkage disequilibrium with ‘nearby’ SNPs (eg SNPs 9 and 10). Again, this somewhat attenuates the problem of multiple comparisons, but does not eliminate it. One Back results remained significant after control of false discovery rate. We also note that it is probably necessary to sample a given cognitive domain with multiple instruments, given psychometric differences among tasks, cohort effects, and an incomplete understanding of the neurobiological effects of the gene of interest. This being said, the problem of multiple comparisons is real and it remains possible that some of the results presented here are spurious. Therefore, they should be accepted with caution until they are replicated in an independent cohort. The pattern of results also supports prior arguments that penetrance may be greater for cognitive phenotypes than clinical diagnosis per se because information processing may be more directly related to genetically driven variants in neuronal signaling.
In conclusion, we have provided evidence that select SNP variations in the G72 gene region increase risk of cognitive impairment in schizophrenia. The nature of this impairment would not be inconsistent with findings from a variety of studies of NMDA-based signaling cascades in excitatory neurotransmission. In particular, attention/working memory, and episodic memory functions became progressively more compromised with increased ‘risk allele load.’ Our results thus extend findings for a role for G72 in serious neuropsychiatric conditions by suggesting that a mutation in G72 may be particularly penetrant at the level of brain information processing implicated in cognitive impairments.
References
Addington AM, Gornick M, Sporn AL, Gogtay N, Greeenstein D, Lenane M et al (2004). Polymorphisms in the 13q33.2 gene G72/G30 are associated with childhood-onset schizophrenia and psychosis not otherwise specified. Biol Psychiatry 55: 976–980.
Brett M (2004). www.mrc-cbu.cam.ac.uk/Imaging/mnispace.htmlCambridge, England.
Burggren AC, Small GW, Sabb FW, Bookheimer SY (2002). Specificity of brain activation patterns in people at genetic risk for Alzheimer disease. Am J Geriatr Psychiatry 10: 44–51.
Callicott JH, Bertolino A, Mattay VS, Langheim FJ, Duyn J, Coppola R et al (2000). Physiological dysfunction of the dorsolateral prefrontal cortex in schizophrenia revisited. Cereb Cortex 10: 1078–1092.
Chen YS, Akula N, Detera-Wadleigh SD (2004). Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Molec Psychiatry 9: 87–92.
Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H et al (2002). Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA 99: 13675–13680.
Clayton D (2001). TRANSMIT v. 2.5. Cambridge, England.
Cloninger CR, Kaufmann CA, Faraone SV, Malaspina D, Svrakic DM, Harkavy-Friedman J et al (1998). Genome-wide search for schizophrenia susceptibility loci: the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet 81: 275–281.
Cox NJ (2002). Calpain 10 and genetics of type 2 diabetes. Curr Diab Rep 2: 186–190.
Deakin JF, Slater P, Simpson MD, Gilchrist AC, Skan WJ, Royston MC et al (1989). Frontal cortical and left temporal glutamatergic dysfunction in schizophrenia. J Neurochem 52: 1781–1786.
Doniger GM, Silipo G, Rabinowicz EF, Snodgrass JG, Javitt DC (2001). Impaired sensory processing as a basis for object-recognition deficits in schizophrenia. Am J Psychiatry 158: 1818–1826.
Egan M, Goldberg TE, Gscheidle T, Weirich M, Rawlings R, Bigelow L et al (2001a). Relative risk for cognitive impairments in siblings of patients with schizophrenia. Biol Psychiatry 50: 98–107.
Egan MF, Goldberg TE, Kojima M, Callicott J, Kolachan BS, Bertolino A et al (2003). The BDNF val66met polymorphism affects activity dependent secretion of BDNF and human memory and hippocampal function. Cell 112: 257–269.
Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE et al (2001b). Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 98: 6917–6922.
Elvevaag B, Weinberger DR, Suter J, Goldberg TE (2000). The Continuous Performance Test and schizophrenia: a test of stimulus response compatibility, working memory, or response readiness? Am J Psychiatry 157: 772–780.
Gabrieli JD, Preston AR (2003). Visualizing genetic influences on human brain function. Cell 112: 144–145.
Gainetdinov RR, Mohn AR, Caron MG (2001). Genetic animal models: focus on schizophrenia. Trends Neurosci 24: 527–533.
Giedd JN, Vaituzis AC, Hamburger SD, Lange N, Rajapakse JC, Kaysen D et al (1996). Quantitative MRI of the temporal lobe, amygdale, and hippocampus in normal human development: ages 4–18 years. J Comp Neurol 366: 223–230.
Goff DC, Coyle JT (2001). The emerging role of glutamate in the pathophysiology and treatment of schizophrenia. Am J Psychiatry 158: 1367–1377.
Goldberg TE, Egan MF, Gsceidle T, Weickert T, Coppola RC, Kolachana BS et al (2003). Executive subprocesses in working memory: Relationships with COMT Val/Met genotype and genetic risk for schizophrenia. Arch Gen Psychiatry 60: 889–896.
Goldberg TE, Green MF (2002). Neurocognitive functioning in patients with schizophrenia: An overview. In Davis KL (ed). Psychopharmacology: The Fifth Generation of Progress. Raven Press: New York. pp 657–669.
Goldberg TE, Ragland DR, Gold J, Bigelow LB, Torrey EF, Weinberger DR (1990). Neuropsychological assessment of monozygotic twins discordant for schizophrenia. Arch Gen Psychiatry 47: 1066–1072.
Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF et al (2003). Brain derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci 23: 6690–6694.
Hariri AR, Weinberger DR (2003). Imaging genomics. Br Med Bull 65: 237–248.
Harrison PJ, Owen MJ (2003). Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet 361: 417–419.
Hattori E, Chunyu L, Badner JA, Bonner TI, Christian SL, Maheshwari M et al (2003). Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet 72: 1131–1140.
Jentsch JD, Roth RH (1999). The neuropsychopharmacology of phencyclidine: from NMDA receptor hypofunction to the dopamine hypothesis of schizophrenia. Neuropsychopharmacology 20: 201–225.
Kaufman AS (1990). Assessing Adolescent and Adult Intelligence. Allyn Bacon: New York, 753 pp.
Krystal JH, Anand A, Moghaddam B (2002). Effects of NMDA receptor antagonists: implications for the pathology of schizophrenia. Arch Gen Psychiatry 59: 663–664.
Krystal JH, Bennett A, Abi-Saab D, Belger A, Karper LP, D'Souza DC et al (2000). Dissociation of ketamine effects on rule acquisition and rule implementation: possible relevance to NMDA receptor contributions to executive cognitive functions. Biol Psychiatry 15: 137–143.
Krystal JH, Karper LP, Bennett A, D'Souza DC, Abi-Dargham A, Morrissey K et al (1998). Interactive effects of subanesthetic ketamine and subhypnotic lorazepam in humans. Biol Psychiatry 135: 213–229.
Krystal JH, Karper LP, Seibyl JP, Freeman GK, Delaney R, Bremner JD et al (1994). Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch Gen Psychiatry 51: 199–214.
Lang PJ, Bradley MM, Cuthbert BN (1997). International Affective Picture System (IAPS): Technical Manual and Affective Ratings. NIMH Center for the Study of Emotion and Attention Gainesville, FL.
Lin Y, Fossom LH, Skolnick P, Long JB (1998). Sustained exposure to a glycine recpetor partial agonist differentially alters NMDA receptor agonist and antagonist potencies of cultured spinal cord neurons. Eur J Pharmacol 356: 255–260.
Littell RC, Milliken GA, Stroup WW, Wolfinger RD (1996). SAS System for Mixed Models. Carey, NC.
Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN (2003). Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33: 177–182.
Malhotra AK, Pinals DA, Weingartner H, Sirocco K, Missar CD, Pickar D et al (1996). NMDA receptor function and human cognition: the effects of ketamine in healthy volunteers. Neuropsychopharmacology 14: 301–307.
Mattay VS, Tessitore A, Callicott JH, Bertolino A, Goldberg TE, Chase TN et al (2002). Dopaminergic modulation of cortical function in patients with Parkinson's disease. Ann Neurol 51: 156–164.
Meador-Woodruff JH, Healy DJ (2000). Glutamate receptor expression in schizophrenic brain. Brain Res Brain Res Rev 31: 288–294.
Pittenger C, Kandel ER (2003). In search of general mechanisms for long-lasting plasticity: Aplysia and the hippocampus. Philos Trans R Soc Lond B Biol Sci 358: 757–763.
Purcell S (2003). Statgen Genetic Power Calculator Module software. Cambridge, England.
Schroeder CE, Lindsley RW, Specht C, Marcovici A, Smiley JF, Javitt DC (2001). Somatosensory input to auditory association cortex in the macaque monkey. J Neurophysiol 85: 1322–1327.
Schumacher J, Jamra R, Freudenberg J (2004). Examination of G72 and d-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Molec Psychiatry 9: 203–207.
Talaraich J, Tournoux P (1988). Co-planar Stereotaxic Atlas of the Human Brain. Thieme Medical Publishers: New York, 122 pp.
Tsai G, Coyle JT (2002). Glutamatergic mechanisms in schizophrenia. Ann Rev Pharmacol Toxicol 42: 165–179.
Tsai G, Passani LA, Slusher BS, Carter R, Baer L, Kleinman JE et al (1995). Abnormal excitatory neurotransmitter metabolism in schizophrenic brains. Arch Gen Psychiatry 52: 829–836.
Weickert T, Egan MF, Weinberger DR, Goldberg TE (2000). Preserved and compromised intellect in schizophrenia: Neurocognitive implications. Arch Gen Psychiatry 57: 907–913.
Wellcome Department of Cognitive Neurology (2000). SPM99. Cambridge, England.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Goldberg, T., Straub, R., Callicott, J. et al. The G72/G30 Gene Complex and Cognitive Abnormalities in Schizophrenia. Neuropsychopharmacol 31, 2022–2032 (2006). https://doi.org/10.1038/sj.npp.1301049
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.npp.1301049
Keywords
This article is cited by
-
Effect of DAOA genetic variation on white matter alteration in corpus callosum in patients with first-episode schizophrenia
Brain Imaging and Behavior (2021)
-
The role of trauma experiences, personality traits, and genotype in maintaining posttraumatic stress disorder symptoms among child survivors of the Wenchuan earthquake
BMC Psychiatry (2020)
-
G72 primate-specific gene: a still enigmatic element in psychiatric disorders
Cellular and Molecular Life Sciences (2016)
-
Identification of the Mitochondrial MSRB2 as a Binding Partner of LG72
Cellular and Molecular Neurobiology (2014)
-
d-amino acid oxidase activator gene (DAOA) variation affects cerebrospinal fluid homovanillic acid concentrations in healthy Caucasians
European Archives of Psychiatry and Clinical Neuroscience (2012)