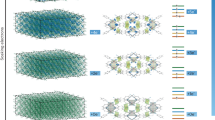
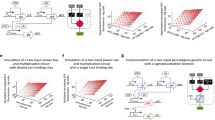
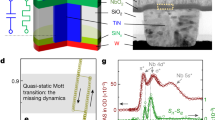
Abstract
Modern computers operate at enormous speeds—capable of executing in excess of 1013 instructions per second—but their sequential approach to processing, by which logical operations are performed one after another, has remained unchanged since the 1950s. In contrast, although individual neurons of the human brain fire at around just 103 times per second, the simultaneous collective action of millions of neurons enables them to complete certain tasks more efficiently than even the fastest supercomputer. Here we demonstrate an assembly of molecular switches that simultaneously interact to perform a variety of computational tasks including conventional digital logic, calculating Voronoi diagrams, and simulating natural phenomena such as heat diffusion and cancer growth. As well as representing a conceptual shift from serial-processing with static architectures, our parallel, dynamically reconfigurable approach could provide a means to solve otherwise intractable computational problems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Higuchi, T. et al. Proc. of ICEC 187–192 (IEEE, 1997).
Neumann, J. V. in The Theory of Self-Reproducing Automata (ed. Burks, A. W.) (Univ. Illinois Press, 1966).
Wolfram, S. A New Kind of Science 223–848 (Wolfram Media Inc., 2002).
Crutchfeld, J. P., Mitchell, M. & Das, R. in Evolutionary Dynamics Exploring the Interplay of Selection, Neutrality, Accident, and Function (eds Crutchfield, J. P. & Schuster, P. K.) (Oxford Univ. Press, 2002).
Hopfield, J. J. & Tank, D. W. Collective computation in neuron like circuits. Sci. Am. 255, 104–114 (1987).
Adamatzky, A. & Teuscher, C. (eds) From Utopian to Genuine Unconventional Computers (Uniliver Press, 2006).
Silva, A. P. & de Uchiyama, S. Molecular logic and computing. Nature Nanotech. 2, 399–410 (2007).
Jones, R. Computing with molecules. Nature Nanotech. 4, 207 (2009).
Credi, A. Monolayers with an IQ. Nature Nanotech. 3, 529–530 (2008).
Lent, C. S., Isaksen, B. & Lieberman, M. Molecular quantum-dot cellular automata. J. Am. Chem. Soc. 125, 1056–1063 (2003).
Orlov, A. O., Amlani, I., Bernstein, G. H., Lent, C. S. & Snider, G. L. Realization of a functional cell for quantum-dot. Cell. Automata Sci. 277, 928–930 (1997).
Qi, H. et al. Molecular quantum cellular automata cells. Electric field driven switching of a silicon surface bound array of vertically oriented two-dot molecular quantum cellular automata. J. Am. Chem. Soc. 125, 15250–15259 (2003).
Imre, A. et al. Majority logic gate for magnetic quantum-dot. Cell. Automata Sci. 311, 205–208 (2006).
Chen, J., Reed, M. A., Rawlett, A. M. & Tour, J. M. Large On–Off ratios and negative differential resistance in a molecular electronic device. Science 286, 1550–1552 (1999).
Pease, A. R. et al. Switching devices based on interlocked molecules. Acc. Chem. Res. 34, 433–444 (2001).
Pati, R., McCLain, M. & Bandyopadhyay, A. Origin of negative differential resistance in strongly coupled molecular junctions. Phys. Rev. Lett. 100, 246801 (2008).
Bandyopadhyay, A., Miki, K. & Wakayama, Y. Writing and erasing information in multilevel logic systems of a single molecule using scanning tunneling microscope (STM). Appl. Phys. Lett. 89, 243507 (2006).
Bandyopadhyay, A. & Acharya, S. A 16-bit parallel processing in a molecular assembly. Proc. Natl Acad. Sci. USA 105, 3668–3672 (2008).
Kirakosian, A., Comstock, M. J., Cho, J. & Crommie, M. F. Molecular commensurability with a surface reconstruction: STM study of azobenzene on Au(111). Phys. Rev. B 71, 113409 (2005).
Cunha, F. & Tao, N. J. Surface charge induced order–disorder transition in an organic monolayer. Phys. Rev. Lett. 75, 2376–2379 (1995).
Onsagar, L. The effect of shape on the interaction of colloidal particles. N.Y. Acad. Sci. 51, 627 (1949).
Müller, T., Werblowsky, T. L., Florio, G. M., Berne, B. J. & Flynn, G. W. Ultra-high vacuum scanning tunneling microscopy and theoretical studies of 1-halohexane monolayers on graphite. Proc. Natl Acad. Sci. USA 102, 5315–5322 (2005).
Dri, C., Peters, M. V., Schwarz, J., Hecht, S. & Grill, L. Spatial periodicity in molecular switching. Nature Nanotech. 3, 649–653 (2008).
Toffoli, T. Cellular automata as an alternative to (rather than an approximation of ) differential equations in modeling. Physica D: Nonlinear Phenom. 10, 117–127 (1984).
Wolfram, S. Method and apparatus for simulating systems described by partial differential equations. US patent number 4,809,202 (1989).
Nowak, M. A. Evolutionary Dynamics: Exploring the Equations of Life (The Belknap Press of Harvard Univ. Press, 2006).
Benenson, Y. et al. Programmable and autonomous computing machine made of biomolecules. Nature 414, 430–434 (2001).
Hjelmfelt, A., Weinberger, E. D. & Ross, J. Chemical implementation of neural networks and Turing machines. Proc. Natl Acad. Sci. USA 8, 10983–10987 (1991).
Ruben, A. J. & Landweber, L. F. The past, present and future of molecular computing. Nature Rev. Mol. Cell Biol. 1, 69–72 (2000).
Landauer, R. Dissipation and noise immunity in computation and communication. Nature 335, 779–784 (1988).
Fredkin, E. & Toffoli, T. Conservative logic. Int. J. Theor. Phys. 21, 219–253 (1982).
Jiménez Morales, F., Crutchfield, J. P. & Mitchell, M. Evolving two-dimensional cellular automata to perform density classification: A report on work in progress. Parallel Comput. 27, 571–585 (2001).
Korobov, A. Discrete versus continual description of solid state reaction dynamics from the angle of meaningful. Simul. Discrete Dynam. Nature Soc. 4, 165–179 (2000).
Gaylord, R. J. & Nishidate, K. Modeling Nature (Springer, 1996).
Acknowledgements
Authors acknowledge H. Hossainkhani, Y. Wakayama, J. Rampe, M. McClain, W. Cantrel and J. Liebescheutz for discussion. The work is partially funded by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan during 2005–2008 and Grants in Aid for Young Scientists (A) for 2009–2011, Grant number 21681015. R.P. acknowledges National Science Foundation (NSF) Award number ECCS-0643420.
Author information
Authors and Affiliations
Contributions
A.B. designed the research; A.B. did the experiment; A.B. developed the CA simulator; R.P., A.B. and S.S. did the theoretical studies; A.B., R.P., S.S. and F.P. analysed the data; and A.B., R.P., F.P. and S.S. wrote the paper together; D.F. reviewed the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 717 kb)
Supplementary Movie
Supplementary Movie 1 (WMV 12229 kb)
Supplementary Movie
Supplementary Movie 2 (WMV 1451 kb)
Supplementary Movie
Supplementary Movie 3 (WMV 1965 kb)
Supplementary Movie
Supplementary Movie 4 (WMV 10441 kb)
Supplementary Movie
Supplementary Movie 5 (WMV 2752 kb)
Rights and permissions
About this article
Cite this article
Bandyopadhyay, A., Pati, R., Sahu, S. et al. Massively parallel computing on an organic molecular layer. Nature Phys 6, 369–375 (2010). https://doi.org/10.1038/nphys1636
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nphys1636
This article is cited by
-
On-chip photonic decision maker using spontaneous mode switching in a ring laser
Scientific Reports (2019)
-
The End of Moore’s Law: Opportunities for Natural Computing?
New Generation Computing (2017)
-
Charge and spin transport in single and packed ruthenium-terpyridine molecular devices: Insight from first-principles calculations
Scientific Reports (2016)
-
Simple and efficient local codes for distributed stable network construction
Distributed Computing (2016)
-
An organic jelly made fractal logic gate with an infinite truth table
Scientific Reports (2015)