Abstract
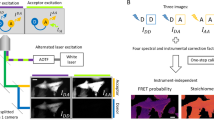
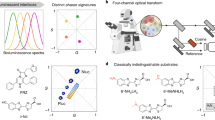
Resonant energy transfer from an optically excited donor molecule to a non-excited acceptor molecule residing nearby is widely used to detect molecular interactions in living cells. To date, resonant energy transfer has been used to obtain stoichiometric information, such as the number of proteins forming a complex, for a handful of proteins, but only after performing sequential scans of the emission wavelengths, excitation wavelengths, or sometimes both. During this lengthy process of measurement, the molecular makeup of a cellular region may change, limiting the applicability of resonant energy transfer to the determination of cellular averages. Here, we demonstrate a method for the determination of protein complex size, configuration, and spatial distribution in single living cells. It relies on a spectrally resolved two-photon microscope, a simple but competent theory, and a judicious selection of fluorescent tags. This approach eventually may lead to tracking the dynamics of individual molecular complexes inside living cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Carriba, P. et al. Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nature Methods 5, 727–733 (2008).
Coulon, V. et al. Subcellular imaging of dynamic protein interactions by bioluminescence resonance energy transfer. Biophys. J. 94, 1001–1009 (2008).
Maurel, D. et al. Cell-surface protein–protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nature Methods 5, 561–567 (2008).
Shyu, Y. J., Suarez, C. D. & Hu, C. D. Visualization of AP-1 NF-κB ternary complexes in living cells by using a BiFC-based FRET. Proc. Natl Acad. Sci. USA 105, 151–156 (2008).
Spriet, C. et al. Enhanced FRET contrast in lifetime imaging. Cytometry A 73, 745–753 (2008).
Chen, Y., Mauldin, J. P., Day, R. N. & Periasamy, A. Characterization of spectral FRET imaging microscopy for monitoring nuclear protein interactions. J. Microsc. 228, 139–152 (2007).
Xu, X. et al. Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc. Natl Acad. Sci. USA 104, 10264–10269 (2007).
Demarco, I. A., Periasamy, A., Booker, C. F. & Day, R. N. Monitoring dynamic protein interactions with photoquenching FRET. Nature Methods 3, 519–524 (2006).
Meyer, B. H. et al. FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells. Proc. Natl Acad. Sci. USA 103, 2138–2143 (2006).
Pfleger, K. D. & Eidne, K. A. Illuminating insights into protein–protein interactions using bioluminescence resonance energy transfer (BRET). Nature Methods 3, 165–174 (2006).
Raicu, V., Jansma, D. B., Miller, R. J. & Friesen, J. D. Protein interaction quantified in vivo by spectrally resolved fluorescence resonance energy transfer. Biochem. J. 385, 265–277 (2005).
Wallrabe, H. & Periasamy, A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16, 19–27 (2005).
An, S. J. & Almers, W. Tracking SNARE complex formation in live endocrine cells. Science 306, 1042–1046 (2004).
Chen, Y. & Periasamy, A. Characterization of two-photon excitation fluorescence lifetime imaging microscopy for protein localization. Microscopy Res. Tech. 63, 72–80 (2004).
Lippincott-Schwartz, J. & Patterson, G. H. Development and use of fluorescent protein markers in living cells. Science 300, 87–91 (2003).
Raicu, V. & Popescu, A. I. Integrated Molecular and Cellular Biophysics (Springer, 2008).
Lakowicz, J. R. Principles of Fluorescence Spectroscopy (Springer, 2006).
Selvin, P. R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7, 730–734 (2000).
Clegg, R. M. in Fluorescence Imaging Spectroscopy and Microscopy (eds Wang, X. F. & Herman, B.) (John Wiley & Sons, 1996).
Raicu, V. Efficiency of resonance energy transfer in homo-oligomeric complexes of proteins. J. Biol. Phys. 33, 109–127 (2007).
Song, L., Hennink, E. J., Young, I. T. & Tanke, H. J. Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophys. J. 68, 2588–2600 (1995).
Kubitscheck, U., Schweitzer-Stenner, R., Arndt-Jovin, D. J., Jovin, T. M. & Pecht, I. Distribution of type I Fc-receptors on the surface of mast cells probed by fluorescence resonance energy transfer. Biophys. J. 64, 110–120 (1993).
Wlodarczyk, J. et al. Analysis of FRET signals in the presence of free donors and acceptors. Biophys. J. 94, 986–1000 (2008).
Neher, R. A. & Neher, E. Applying spectral fingerprinting to the analysis of FRET images. Microsc. Res. Tech. 64, 185–195 (2004).
Zimmermann, T., Rietdorf, J., Girod, A., Georget, V. & Pepperkok, R. Spectral imaging and linear un-mixing enables improved FRET efficiency with a novel GFP2–YFP FRET pair. FEBS Lett. 531, 245–249 (2002).
Lee, N. K. et al. Three-color alternating-laser excitation of single molecules: monitoring multiple interactions and distances. Biophys. J. 92, 303–312 (2007).
Eggeling, C., Kask, P., Winkler, D. & Jager, S. Rapid analysis of Forster resonance energy transfer by two-color global fluorescence correlation spectroscopy: trypsin proteinase reaction. Biophys. J. 89, 605–618 (2005).
Angers, S. et al. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl Acad. Sci. USA 97, 3684–3689 (2000).
Xu, Y., Piston, D. W. & Johnson, C. H. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl Acad. Sci. USA 96, 151–156 (1999).
Hoshino, H., Nakajima, Y. & Ohmiya, Y. Luciferase-YFP fusion tag with enhanced emission for single-cell luminescence imaging. Nature Methods 4, 637–639 (2007).
Raicu, V. & Fung, R. US patent application 11/904,860 (2007).
Zipfel, W. R., Williams, R. M. & Webb, W. W. Nonlinear magic: multiphoton microscopy in the biosciences. Nature Biotechnol. 21, 1369–1377 (2003).
Denk, W., Strickler, J. H. & Webb, W. W. Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990).
Kumazaki, S. et al. A line-scanning semi-confocal multi-photon fluorescence microscope with a simultaneous broadband spectral acquisition and its application to the study of the thylakoid membrane of a cyanobacterium Anabaena PCC7120. J. Microsc. 228, 240–254 (2007).
Raicu, V., Fung, R., Melnichuk, M., Chaturvedi, A. & Gillman, D. in Proc. SPIE (eds Periasamy, A. & So, P. T. C.) 6442 M6441–6442 M6445 (SPIE, 2007).
Weiner, A. M. Femtosecond pulse shaping using spatial light modulators. Rev. Sci. Instrum. 71, 1929–1960 (2000).
Raicu, V. et al. in Proc. SPIE (eds Periasamy, A. & So, P. T. C.) 686018 (SPIE, 2008).
Dela Cruz, J. M., Lozovoy, V. V. & Dantus, M. Coherent control improves biomedical imaging with ultrashort shaped pulses. J. Photochem. Photobiol. A: Chemistry 180, 307–313 (2006).
Overton, M. C., Chinault, S. L. & Blumer, K. J. Oligomerization of G-protein-coupled receptors: Lessons from the yeast Saccharomyces cerevisiae. Eukaryotic Cell 4, 1963–1970 (2005).
Duncan, R. R., Bergmann, A., Cousin, M. A., Apps, D. K. & Shipston, M. J. Multi-dimensional time-correlated single photon counting (TCSPC) fluorescence lifetime imaging microscopy (FLIM) to detect FRET in cells. J. Microsc. 215, 1–12 (2004).
James, J. R., Oliveira, M. I., Carmo, A. M., Iaboni, A. & Davis, S. J. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nature Methods 3, 1001–1006 (2006).
Kenworthy, A. K. & Edidin, M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 Å using imaging fluorescence resonance energy transfer. J. Cell Biol. 142, 69–84 (1998).
Wolber, P. K. & Hudson, B. S. An analytic solution to the Forster Energy Transfer problem in two dimensions. Biophys. J. 28, 197–210 (1979).
Goin, J. C. & Nathanson, N. M. Quantitative analysis of muscarinic acetylcholine receptor homo- and heterodimerization in live cells: regulation of receptor down-regulation by heterodimerization. J. Biol. Chem. 281, 5416–5425 (2006).
Mercier, J. F., Salahpour, A., Angers, S., Breit, A. & Bouvier, M. Quantitative assessment of beta 1- and beta 2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J. Biol. Chem. 277, 44925–44931 (2002).
Kim, K. H., Buehler, C. & So, P. T. C. High-speed, two-photon scanning microscope. Appl. Opt. 38, 6004–6009 (1999).
Raymond, C. K., Sims, E. H. & Olson, M. V. Linker-mediated recombinational subcloning of large DNA fragments using yeast. Genome Res. 12, 190–197 (2002).
Kurjan, J. Pheromone response in yeast. Annu. Rev. Biochem. 61, 1097–1129 (1992).
Stefan, C. J. & Blumer, K. J. A syntaxin homolog encoded by VAM3 mediates down-regulation of a yeast G protein-coupled receptor. J. Biol. Chem. 274, 1835–1841 (1999).
Acknowledgements
This work was supported by a grant from the Wisconsin Institute for Biomedical and Health Technology (grant no. W620 to V.R.), seed funds from the UWM Research Growth Initiative (grant no. X014 to V.R.), and a grant from Canadian Institutes of Health Research (grant no. MOP43990 to J.W.W.). We thank D. Gillman for useful suggestions regarding the computer routines for instrument control, M. J. Woodside for his assistance with the lifetime imaging, and S. Angers for very useful discussions on BRET. We thank K. J. Blumer for providing YFP.
Author information
Authors and Affiliations
Contributions
V.R. coordinated the project, designed the TPM, built the optical set-up, developed the theory and the algorithms for image reconstruction and data analysis, analysed data, and wrote the paper. M.S., S.R. and M.F. carried out experiments using the TPM and analysed data. R.F. wrote computer codes for hardware control and for image reconstruction. M.M. interfaced the scanners with the computer and built an earlier version of the optical set-up. D.B.J. made the genetic constructs. L.P. and J.W.W. determined the RET efficiency using FLIM. D.K.S. participated in theoretical modelling. All authors contributed to manuscript editing.
Corresponding authors
Supplementary information
Rights and permissions
About this article
Cite this article
Raicu, V., Stoneman, M., Fung, R. et al. Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nature Photon 3, 107–113 (2009). https://doi.org/10.1038/nphoton.2008.291
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nphoton.2008.291
This article is cited by
-
Suberin, the hallmark constituent of bark, identified in a 45-million-year-old monkeyhair tree (Coumoxylon hartigii) from Geiseltal, Germany
Scientific Reports (2024)
-
Two-photon excitation fluorescence microspectroscopy protocols for examining fluorophores in fossil plants
Communications Biology (2024)
-
A general method to quantify ligand-driven oligomerization from fluorescence-based images
Nature Methods (2019)
-
The EphA2 receptor is activated through induction of distinct, ligand-dependent oligomeric structures
Communications Biology (2018)
-
Metschnikowia mating genomics
Antonie van Leeuwenhoek (2018)