Abstract
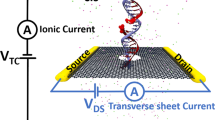
Solid-state nanopores can act as single-molecule sensors and could potentially be used to rapidly sequence DNA molecules. However, nanopores are typically fabricated in insulating membranes that are as thick as 15 bases, which makes it difficult for the devices to read individual bases. Graphene is only 0.335 nm thick (equivalent to the spacing between two bases in a DNA chain) and could therefore provide a suitable membrane for sequencing applications. Here, we show that a solid-state nanopore can be integrated with a graphene nanoribbon transistor to create a sensor for DNA translocation. As DNA molecules move through the pore, the device can simultaneously measure drops in ionic current and changes in local voltage in the transistor, which can both be used to detect the molecules. We examine the correlation between these two signals and use the ionic current measurements as a real-time control of the graphene-based sensing device.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Li, J. et al. Ion-beam sculpting at nanometre length scales. Nature 412, 166–169 (2001).
Li, J. L., Gershow, M., Stein, D., Brandin, E. & Golovchenko, J. A. DNA molecules and configurations in a solid-state nanopore microscope. Nature Mater. 2, 611–615 (2003).
Fologea, D. et al. Detecting single stranded DNA with a solid state nanopore. Nano Lett. 5, 1905–1909 (2005).
Storm, A. J., Chen, J. H., Zandbergen, H. W. & Dekker, C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys. Rev. E 71, 051903 (2005).
Dekker, C. et al. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 6, 89–95 (2006).
Skinner, G. M., van den Hout, M., Broekmans, O., Dekker, C. & Dekker, N. H. Distinguishing single- and double-stranded nucleic acid molecules using solid-state nanopores. Nano Lett. 9, 2953–2960 (2009).
Van den Hout, M., Krudde, V., Janssen, X. J. A. & Dekker, N. H. Distinguishable populations report on the interactions of single DNA molecules with solid-state nanopores. Biophys. J. 99, 3840–3848 (2010).
Wanunu, M. et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nature Nanotech. 5, 807–814 (2010).
Keyser, U. F. et al. Direct force measurements on DNA in a solid-state nanopore. Nature Phys. 2, 473–477 (2006).
Trepagnier, E. H., Radenovic, A., Sivak, D., Geissler, P. & Liphardt, J. Controlling DNA capture and propagation through artificial nanopores. Nano Lett. 7, 2824–2830 (2007).
Dekker, C. Solid-state nanopores. Nature Nanotech. 2, 209–215 (2007).
Peng, H. B. & Ling, X. S. S. Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotechnology 20, 185101 (2009).
Taniguchi, M., Tsutsui, M., Yokota, K. & Kawai, T. Fabrication of the gating nanopore device. Appl. Phys. Lett. 95, 123701 (2009).
McNally, B. et al. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 10, 2237–2244 (2010).
Tsutsui, M. et al. Single-molecule sensing electrode embedded in-plane nanopore. Sci. Rep. 1, 46 (2011).
Xie, P., Xiong, Q. H., Fang, Y., Qing, Q. & Lieber, C. M. Local electrical potential detection of DNA by nanowire–nanopore sensors. Nature Nanotech. 7, 119–125 (2012).
Geim, A. K. Graphene: status and prospects. Science 324, 1530–1534 (2009).
Garaj, S. et al. Graphene as a subnanometre trans-electrode membrane. Nature 467, 190–193 (2010).
Merchant, C. A. et al. DNA translocation through graphene nanopores. Nano Lett. 10, 2915–2921 (2010).
Schneider, G. F. et al. DNA translocation through graphene nanopores. Nano Lett. 10, 3163–3167 (2010).
Lagerqvist, J., Zwolak, M. & Di Ventra, M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 6, 779–782 (2006).
Postma, H. W. Rapid sequencing of individual DNA molecules in graphene nanogaps. Nano Lett. 10, 420–425 (2010).
Min, S. K., Kim, W. Y., Cho, Y. & Kim, K. S. Fast DNA sequencing with a graphene-based nanochannel device. Nature Nanotech. 6, 162–165 (2011).
Ivanov, A. P. et al. DNA tunneling detector embedded in a nanopore. Nano Lett. 11, 279–285 (2011).
Storm, A. J., Chen, J. H., Ling, X. S., Zandbergen, H. W. & Dekker, C. Fabrication of solid-state nanopores with single-nanometre precision. Nature Mater. 2, 537–540 (2003).
Ang, P. K., Chen, W., Wee, A. T. S. & Loh, K. P. Solution-gated epitaxial graphene as pH sensor. J. Am. Chem. Soc. 130, 14392–14393 (2008).
Smeets, R. M. M., Dekker, N. H. & Dekker, C. Low-frequency noise in solid-state nanopores. Nanotechnology 20, 095501 (2009).
Kuan, A. T. & Golovchenko, J. A. Nanometer-thin solid-state nanopores by cold ion beam sculpting. Appl. Phys. Lett. 100, 213104 (2012).
Dimitrov, V. et al. Nanopores in solid-state membranes engineered for single molecule detection. Nanotechnology 21, 065502 (2010).
Venkatesan, B. M., Shah, A. B., Zuo, J. M. & Bashir, R. DNA sensing using nanocrystalline surface-enhanced Al2O3 nanopore sensors. Adv. Funct. Mater. 20, 1266–1275 (2010).
Raillon, C. et al. Nanopore detection of single molecule RNAP–DNA transcription complex. Nano Lett. 12, 1157–1164 (2012).
Raillon, C., Granjon, P., Graf, M., Steinbock, L. J. & Radenovic, A. Fast and automatic processing of multi-level events in nanopore translocation experiments. Nanoscale 4, 4916–4924 (2012).
Van Dorp, S., Keyser, U. F., Dekker, N. H., Dekker, C. & Lemay, S. G. Origin of the electrophoretic force on DNA in solid-state nanopores. Nature Phys. 5, 347–351 (2009).
Heller, I. et al. Comparing the weak and strong gate-coupling regimes for nanotube and graphene transistors. Phys. Status Solidi 3, 190–192 (2009).
Harrell, C. C. et al. Resistive-pulse DNA detection with a conical nanopore sensor. Langmuir 22, 10837–10843 (2006).
Wanunu, M., Morrison, W., Rabin, Y., Grosberg, A. Y. & Meller, A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nature Nanotech. 5, 160–165 (2010).
Novoselov, K. S. et al. Two-dimensional atomic crystals. Proc. Natl Acad. Sci. USA 102, 10451–10453 (2005).
Radisavljevic, B., Radenovic, A., Brivio, J., Giacometti, V. & Kis, A. Single-layer MoS2 transistors. Nature Nanotech. 6, 147–150 (2011).
Blake, P. et al. Making graphene visible. Appl. Phys. Lett. 91, 063124 (2007).
Li, X. S. et al. Large-area synthesis of high-quality and uniform graphene films on copper foils. Science 324, 1312–1314 (2009).
Ye, Y. et al. A simple and scalable graphene patterning method and its application in CdSe nanobelt/graphene Schottky junction solar cells. Nanoscale 3, 1477–1481 (2011).
Vlassarev, D. M. & Golovchenko, J. A. Trapping DNA near a solid-state nanopore. Biophys. J. 103, 352–356 (2012).
Acknowledgements
This work was supported financially by the European Research Council (grant no. 259398, PorABEL: Nanopore Integrated Nanoelectrodes for Biomolecular Manipulation and Sensing). F.T. was partially financed by an FP7 nanoDNA sequencing grant. C.R. was financed by a grant from the Swiss SystemsX.ch initiative (IPhD), evaluated by the Swiss National Science Foundation. M.B. and D.K. were supported by grants from the Swiss National Science Foundation (grants nos 122044 and 135046). The authors thank the Centre Interdisciplinaire de Microscopie Electronique (CIME) at EPFL for access to electron microscopes, and special thanks go to D.T.L. Alexander for providing training and technical assistance with TEM. Device fabrication was partially carried out at the EPFL Center for Micro/Nanotechnology (CMi). The authors thank A. Ionescu and Nanolab (EPFL) for access to Advance Design System (ADS) software and A. Bazigos for help with ADS. Thanks go to V. Russo and C.S. Casari of the Politecnico di Milano for Raman spectroscopy on graphene, P. Granjon (Grenoble Institute of Technology) for help with the crosstalk analysis, and all LBEN laboratory members for reviewing the manuscript.
Author information
Authors and Affiliations
Contributions
F.T., A.K. and A.R. designed the research. F.T. and A.R. designed and built the measurement set-up. F.T. fabricated and characterized devices, performed experiments and analysed the data. K.L. performed experiments and analysed the data. C.R. and M.B. worked on the first generation of devices based on exfoliated graphene. S.K. performed COMSOL modelling. M.T. and D.K. performed CVD graphene growth and transfer. F.T., A.K. and A.R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Information (PDF 3265 kb)
Rights and permissions
About this article
Cite this article
Traversi, F., Raillon, C., Benameur, S. et al. Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nature Nanotech 8, 939–945 (2013). https://doi.org/10.1038/nnano.2013.240
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2013.240
This article is cited by
-
Scalable graphene sensor array for real-time toxins monitoring in flowing water
Nature Communications (2023)
-
Low-dimensionality carbon-based biosensors: the new era of emerging technologies in bioanalytical chemistry
Analytical and Bioanalytical Chemistry (2023)
-
DNA bases detection via MoS2 field effect transistor with a nanopore: first-principles modeling
Analog Integrated Circuits and Signal Processing (2023)
-
DNA sequencing: an overview of solid-state and biological nanopore-based methods
Biophysical Reviews (2022)
-
Localised solid-state nanopore fabrication via controlled breakdown using on-chip electrodes
Nano Research (2022)