Abstract
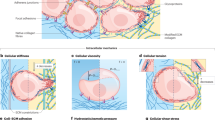
Cancer initiation and progression follow complex molecular and structural changes in the extracellular matrix and cellular architecture of living tissue. However, it remains poorly understood how the transformation from health to malignancy alters the mechanical properties of cells within the tumour microenvironment. Here, we show using an indentation-type atomic force microscope (IT-AFM) that unadulterated human breast biopsies display distinct stiffness profiles. Correlative stiffness maps obtained on normal and benign tissues show uniform stiffness profiles that are characterized by a single distinct peak. In contrast, malignant tissues have a broad distribution resulting from tissue heterogeneity, with a prominent low-stiffness peak representative of cancer cells. Similar findings are seen in specific stages of breast cancer in MMTV-PyMT transgenic mice. Further evidence obtained from the lungs of mice with late-stage tumours shows that migration and metastatic spreading is correlated to the low stiffness of hypoxia-associated cancer cells. Overall, nanomechanical profiling by IT-AFM provides quantitative indicators in the clinical diagnostics of breast cancer with translational significance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ingber, D. E. et al. Cellular tensegrity—exploring how mechanical changes in the cytoskeleton regulate cell-growth, migration, and tissue pattern during morphogenesis. Int. Rev. Cytol. 150, 173–224 (1994).
Park, C. C., Bissell, M. J. & Barcellos-Hoff, M. H. The influence of the microenvironment on the malignant phenotype. Mol. Med. Today 6, 324–329 (2000).
Needham, D. Possible role of cell cycle-dependent morphology, geometry, and mechanical-properties in tumor-cell metastasis. Cell Biophys. 18, 99–121 (1991).
Paszek, M. J. & Weaver, V. M. The tension mounts: mechanics meets morphogenesis and malignancy. J. Mammary Gland Biol. 9, 325–342 (2004).
Kumar, S. & Weaver, V. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metast. Rev. 28, 113–127 (2009).
Kass, L., Erler, J. T., Dembo, M. & Weaver, V. M. Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. Int. J. Biochem. Cell B 39, 1987–1994 (2007).
Butcher, D. T., Alliston, T. & Weaver, V. M. A tense situation: forcing tumour progression. Nature Rev. Cancer 9, 108–122 (2009).
Sinkus, R. et al. High-resolution tensor MR elastography for breast tumour detection. Phys. Med. Biol. 45, 1649–1664 (2000).
Paszek, M. J. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Levental, K. R. et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009).
Rosenbluth, M. J., Lam, W. A. & Fletcher, D. A. Force microscopy of nonadherent cells: a comparison of leukemia cell deformability. Biophys. J. 90, 2994–3003 (2006).
Cross, S. E., Jin, Y. S., Rao, J. & Gimzewski, J. K. Nanomechanical analysis of cells from cancer patients. Nature Nanotech. 2, 780–783 (2007).
Lekka, M. et al. Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur. Biophys. J. Biophy. 28, 312–316 (1999).
Ward, K. A., Li, W. I., Zimmer, S. & Davis, T. Viscoelastic properties of transformed-cells—role in tumor-cell progression and metastasis formation. Biorheology 28, 301–313 (1991).
Lam, W. A., Rosenbluth, M. J. & Fletcher, D. A. Chemotherapy exposure increases leukemia cell stiffness. Blood 109, 3505–3508 (2007).
Guck, J. et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 88, 3689–3698 (2005).
Suresh, S. Biomechanics and biophysics of cancer cells. Acta Mater. 55, 3989–4014 (2007).
Lekka, M. et al. Cancer cell detection in tissue sections using AFM. Arch. Biochem. Biophys. 518, 151–156 (2012).
Weaver, V. M. et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009).
Erler, J. T. & Weaver, V. M. Three-dimensional context regulation of metastasis. Clin. Exp. Metastasis 26, 35–49 (2009).
Weaver, V. M., DuFort, C. C. & Paszek, M. J. Balancing forces: architectural control of mechanotransduction. Nature Rev. Mol. Cell. Biol. 12, 308–319 (2011).
Lin, E. Y. et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 (2003).
Lundin, M., Lundin, J., Helin, H. & Isola, J. A digital atlas of breast histopathology: an application of web based virtual microscopy. J. Clin. Pathol. 57, 1288–1291 (2004).
Weaver, V. M. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Egeblad, M. & Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nature Rev. Cancer 2, 161–174 (2002).
Albrechtsen, R., Nielsen, M., Wewer, U., Engvall, E. & Ruoslahti, E. Basement membrane changes in breast cancer detected by immunohistochemical staining for laminin. Cancer Res. 41, 5076–5081 (1981).
Guy, C. T., Cardiff, R. D. & Muller, W. J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell Biol. 12, 954–961 (1992).
Fantozzi, A. & Christofori, G. Mouse models of breast cancer metastasis. Breast Cancer Res. 8, 212 (2006).
Guppy, M. The hypoxic core: a possible answer to the cancer paradox. Biochem. Biophys. Res. Commun. 299, 676–680 (2002).
Sedwick, C. Valerie Weaver: overcoming cancer's stiff resistance. J. Cell. Biol. 193, 802–803 (2011).
Lopez, J. I., Kang, I., You, W. K., McDonald, D. M. & Weaver, V. M. In situ force mapping of mammary gland transformation. Integr. Biol. (Camb.) 3, 910–921 (2011).
Suresh, S. Biomechanics and biophysics of cancer cells. Acta Biomater. 3, 413–438 (2007).
Alcaraz, J. et al. Collective epithelial cell invasion overcomes mechanical barriers of collagenous extracellular matrix by a narrow tube-like geometry and MMP14-dependent local softening. Integr. Biol. (Camb.) 3, 1153–1166 (2011).
Stolz, M. et al. Early detection of aging cartilage and osteoarthritis in mice and patient samples using atomic force microscopy. Nature Nanotech. 4, 186–192 (2009).
Thomas, A. et al. Real-time elastography—an advanced method of ultrasound: first results in 108 patients with breast lesions. Ultrasound Obst. Gyn. 28, 335–340 (2006).
Burnside, E. S. et al. Differentiating benign from malignant solid breast masses with US strain imaging. Radiology 245, 401–410 (2007).
Xu, H. Y. et al. Axial-shear strain imaging for differentiating benign and malignant breast masses. Ultrasound Med. Biol. 36, 1813–1824 (2010).
Wong, C. C. L. et al. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc. Natl Acad. Sci. USA 108, 16369–16374 (2011).
Erler, J. T. et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 440, 1222–1226 (2006).
Fritsch, A. et al. Are biomechanical changes necessary for tumour progression? Nature Phys. 6, 730–732 (2010).
Wirtz, D., Konstantopoulos, K. & Searson, P. C. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nature Rev. Cancer 11, 512–522 (2011).
Erler, J. T., Jeffrey, S. S. & Giaccia, A. J. Hypoxia promotes invasion and metastasis of breast cancer cells by increasing lysyl oxidase expression. Breast Cancer Res. 7, S57 (2005).
Sader, J. E., Larson, I., Mulvaney, P. & White, L. R. Method for the calibration of atomic-force microscope cantilevers. Rev. Sci. Instrum. 66, 3789–3798 (1995).
Oliver, W. C. & Pharr, G. M. An improved technique for determining hardness and elastic-modulus using load and displacement sensing indentation experiments. J. Mater. Res. 7, 1564–1583 (1992).
Plodinec, M., Loparic, M. & Aebi, U. Atomic force microscopy for biological imaging and mechanical testing across length scales. Cold Spring Harb. Protoc. 2010, pdb top86 (2010).
Loparic, M. et al. Micro- and nanomechanical analysis of articular cartilage by indentation-type atomic force microscopy: validation with a gel-microfiber composite. Biophys. J. 98, 2731–2740 (2010).
Plodinec, M. et al. The nanomechanical properties of rat fibroblasts are modulated by interfering with the vimentin intermediate filament system. J. Struct. Biol. 174, 476–484 (2011).
Acknowledgements
The authors thank U. Mueller for excising tissues from MMTV-PyMT mice, T. Nguyen and P. Hirschmann for technical assistance with histology and IHC, and R. Suetterlin for advice on IHC. B. Bircher is acknowledged for his contribution to AFM data analysis, T. Pfändler for logistic support concerning clinical samples and A. Roulier for help with the drawing in Fig. 1. The authors also thank U. Sauder for SEM sample preparation, D. Mathys for SEM imaging and P. Demougin for RNA extraction. This work is funded by the National Centre of Competence in Research ‘Nanoscale Science’, Swiss National Science Foundation (to C-A.S.), and the Commission for Technology and Innovation (CTI) supporting university–industry partnerships (Project 11977.2 PFNM-NM within the project ARTIDIS ‘Automated and Reliable Tissue Diagnostics’ awarded to R.Y.H.L in partnership with Nanosurf AG). R.Z.D. is supported by Krebsliga Beider Basel (grant no. 22-2010). The laboratory of M.B-A. is supported by the Novartis Research Foundation, the European Research Council (ERC starting grant no. 243211-PTPsBDC), the Swiss Cancer League and the Krebsliga Beider Basel.
Author information
Authors and Affiliations
Contributions
M.P., R.Y.H.L. and C-A.S. conceived the study and designed experiments. M.P., M.L. and R.Y.H.L. developed all customized hardware and software solutions for AFM. M.P. and E.C.O. performed pathohistological and IHC analysis of human and murine tissues. R.Z.D. recruited patients and provided human biopsies. M.P., C.A.M. and P.O. performed AFM experiments. M.P., M.L, C.A.M., J.T.H., P.O. and R.Y.H.L. analysed AFM data. M.B-A. provided MMTV-PyMT mice and was involved in the analysis of murine tissues. M.P., U.A., R.Y.H.L. and C-A.S. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The University of Basel has filed patents on the technology and intellectual property related to this work based on the inventions of M.P., M.L. and R.Y.H.L.
Supplementary information
Supplementary information
Supplementary information (PDF 2849 kb)
Rights and permissions
About this article
Cite this article
Plodinec, M., Loparic, M., Monnier, C. et al. The nanomechanical signature of breast cancer. Nature Nanotech 7, 757–765 (2012). https://doi.org/10.1038/nnano.2012.167
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2012.167
This article is cited by
-
Evidence and therapeutic implications of biomechanically regulated immunosurveillance in cancer and other diseases
Nature Nanotechnology (2024)
-
Optical Elastography for Micropressure Characterization of Zebrafish Embryonic Cardiac Development
Annals of Biomedical Engineering (2024)
-
Cancer stem cells and their niche in cancer progression and therapy
Cancer Cell International (2023)
-
Tumor matrix stiffness provides fertile soil for cancer stem cells
Cancer Cell International (2023)
-
An exploratory study of cell stiffness as a mechanical label-free biomarker across multiple musculoskeletal sarcoma cells
BMC Cancer (2023)