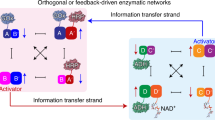
Abstract
The ability of DNA to self-assemble into one-, two- and three-dimensional nanostructures1,2,3,4,5,6,7,8,9,10,11,12,13,14, combined with the precision that is now possible when positioning nanoparticles15,16,17,18,19 or proteins20,21,22,23,24 on DNA scaffolds, provide a promising approach for the self-organization of composite nanostructures25,26,27. Predicting and controlling the functions that emerge in self-organized biomolecular nanostructures is a major challenge in systems biology, and although a number of innovative examples have been reported28,29,30, the emergent properties of systems in which enzymes are coupled together have not been fully explored. Here, we report the self-assembly of a DNA scaffold made of DNA strips that include ‘hinges’ to which biomolecules can be tethered. We attach either two enzymes or a cofactor–enzyme pair to the scaffold, and show that enzyme cascades or cofactor-mediated biocatalysis can proceed effectively; similar processes are not observed in diffusion-controlled homogeneous mixtures of the same components. Furthermore, because the relative position of the two enzymes or the cofactor–enzyme pair is determined by the topology of the DNA scaffold, it is possible to control the reactivity of the system through the design of the individual DNA strips. This method could lead to the self-organization of complex multi-enzyme cascades.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Kallenbach, N. R., Ma, R.-I. & Seeman, N. C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 305, 829–831 (1983).
Mao, C., Sun, W. & Seeman, N. C. Designed two-dimensional DNA Holliday junction arrays visualised by atomic force microscopy. J. Am. Chem. Soc. 121, 5437–5443 (1999).
Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Yang, X., Wenzler, L. A., Qi, J., Li, X. & Seeman, N. C. Ligation of DNA triangles containing double crossover molecules. J. Am. Chem. Soc. 120, 9779–9786 (1998).
Ding, B., Sha, R. & Seeman, N. C. Pseudohexagonal 2D crystals from double crossover cohesion. J. Am. Chem. Soc. 126, 10230–10231 (2004).
Park, S. H., Finkelstein, G. & LaBean, T. H. Stepwise self-assembly of DNA tile lattices using dsDNA bridges. J. Am. Chem. Soc. 130, 40–41 (2008).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Zhang, C., He, Y., Chen, Y., Ribbe, A. E. & Mao, C. Aligning one-dimensional DNA duplexes into two-dimensional crystals. J. Am. Chem. Soc. 129, 14134–14135 (2007).
Chen, J. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Mao, C., Sun, W. & Seeman, N. C. Assembly of Borromean rings from DNA. Nature 386, 137–138 (1997).
Kuzuya, A., Wang, R., Sha, R. & Seeman, N. C. Six-helix and eight-helix DNA nanotubes assembled from half-tubes. Nano Lett. 7, 1757–1763 (2007).
Faisal, A. A. & Sleiman, H. F. Modular access to structurally switchable 3D discrete DNA assemblies. J. Am. Chem. Soc. 129, 13376–13377 (2007).
Simmel, F. C. Three-dimensional nanoconstruction with DNA. Angew. Chem. Int. Ed. 47, 5884–5887 (2008).
Le, J. D. et al. DNA-templated self-assembly of metallic nanocomponents arrays on a surface. Nano Lett. 4, 2343–2347 (2004).
Nykypanchuk, D., Maye, M. M., van der Lelie, D. & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008).
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008).
Sharma, J. et al. DNA-tile-directed self-assembly of quantum dots into two-dimensional nanopatterns. Angew. Chem. Int. Ed. 47, 5157–5159 (2008).
Li, H., Park, S. H., Reif, J. H., LaBean, T. H. & Yan, H. DNA templated self-assembly of protein and nanoparticle linear arrays. J. Am. Chem. Soc. 126, 418–419 (2004).
Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301, 1882–1884 (2003).
Park, S. H. et al. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 5, 729–733 (2005).
Cohen, J. D., Sadowski, J. P. & Dervan, P. B. Addressing single molecules on DNA nanostructures. Angew. Chem. Int. Ed. 46, 7956–7959 (2007).
Liu, Y., Lin, C., Li, H. & Yan, H. Aptamer-directed self-assembly of protein arrays on a DNA nanostructure. Angew. Chem. Int. Ed. 44, 4333–4338 (2005).
Rinker, S., Ke, Y., Liu, Y., Chhabra, R. & Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand–protein binding. Nature Nanotech. 3, 418–422 (2008).
Fruk, L. et al. DNA-directed immobilization of horseradish peroxidase–DNA conjugates on microelectrode arrays: towards electrochemical screening of enzyme libraries. Chem. Eur. J. 13, 5223–5231 (2007).
Cheglakov, Z., Weizmann, Y., Braunschweig, A. B., Wilner, O. I. & Willner, I. Increasing the complexity of periodic protein nanostructures by the rolling-circle-amplified synthesis of aptamers. Angew. Chem. Int. Ed. 47, 126–130 (2008).
Weizmann, Y., Braunschweig, A. B., Wilner, O. I., Cheglakov, Z. & Willner, I. A polycatenated DNA scaffold for the one-step assembly of hierarchical nanostructures. Proc. Natl Acad. Sci. USA 105, 5289–5294 (2008).
Diehl, M. R., Zhang, K., Lee, H. J. & Tirrell, D. A. Engineering cooperativity in biomotor–protein assemblies. Science 311, 1468–1471 (2006).
Bashor, C. J., Helman, N. C., Yan, S. & Lim, W. A. Using engineered scaffold interactions to reshape map kinase pathway signaling dynamics. Science 319, 1539–1543 (2008).
Niemeyer, C. M., Koehler, J. & Wuerdemann, C. DNA-directed assembly of bienzymic complexes from in vivo biotinylated NAD(P)H:FMN oxidoreductase and luciferase. ChemBioChem 3, 242–245 (2002).
Acknowledgements
This research is supported by the Converging Technologies Fund, administered by the Israel Science Foundation. We thank N. Melamed-Book from the Confocal Microscope Unit, Institute of Life Science, The Hebrew University of Jerusalem for experimental assistance.
Author information
Authors and Affiliations
Contributions
O.I.W. designed and performed the experiments, analysed the data and co-wrote the paper. Y.W. and R.G. contributed to the design of the systems and data analysis. O.L. contributed to the data analysis. R.F. participated in data analysis and co-wrote the paper, and I.W. contributed to the design of the systems, data analysis and co-writing of the paper.
Corresponding author
Supplementary information
Supplementary Information
Supplementary Information (PDF 1601 kb)
Rights and permissions
About this article
Cite this article
Wilner, O., Weizmann, Y., Gill, R. et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nature Nanotech 4, 249–254 (2009). https://doi.org/10.1038/nnano.2009.50
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2009.50
This article is cited by
-
Research progress of multi-enzyme complexes based on the design of scaffold protein
Bioresources and Bioprocessing (2023)
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)
-
Spatiotemporal control for integrated catalysis
Nature Reviews Methods Primers (2023)
-
Nucleic acid-based scaffold systems and application in enzyme cascade catalysis
Applied Microbiology and Biotechnology (2023)
-
Trade-offs and design principles in the spatial organization of catalytic particles
Nature Physics (2022)