Abstract
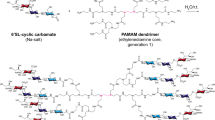
Rapid change1 and zoonotic transmission to humans2 have enhanced the virulence of the influenza A virus (IAV)3. Neutralizing antibodies fail to provide lasting protection from seasonal epidemics1,4. Furthermore, the effectiveness of anti-influenza neuraminidase inhibitors has declined because of drug resistance5. Drugs that can block viral attachment and cell entry independent of antigenic evolution or drug resistance might address these problems. We show that multivalent 6′-sialyllactose-polyamidoamine (6SL–PAMAM) conjugates, when designed to have well-defined ligand valencies and spacings, can effectively inhibit IAV infection. Generation 4 (G4) 6SL–PAMAM conjugates with a spacing of around 3 nm between 6SL ligands (S3–G4) showed the strongest binding to a hemagglutinin trimer (dissociation constant of 1.6 × 10−7 M) and afforded the best inhibition of H1N1 infection. S3–G4 conjugates were resistant to hydrolysis by H1N1 neuraminidase. These conjugates protected 75% of mice from a lethal challenge with H1N1 and prevented weight loss in infected animals. The structure-based design of multivalent nanomaterials, involving modulation of nanoscale backbone structures and number and spacing between ligands, resulted in optimal inhibition of IAV infection. This approach may be broadly applicable for designing effective and enduring therapeutic protection against human or avian influenza viruses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Smith, D. J. et al. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004).
Lam, T. T. et al. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature 522, 102–105 (2015).
Schrauwen, E. J. et al. Determinants of virulence of influenza A virus. Eur. J. Clin. Microbiol. Infect. Dis. 33, 479–490 (2014).
Schrauwen, E. J. & Fouchier, R. A. Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microbes Infect. 3, e9 (2014).
Moscona, A. Global transmission of oseltamivir-resistant influenza. N. Engl. J. Med. 360, 953–956 (2009).
Shi, Y., Wu, Y., Zhang, W., Qi, J. & Gao, G. F. Enabling the ‘host jump’: structural determinants of receptor-binding specificity in influenza A viruses. Nat. Rev. Microbiol. 12, 822–831 (2014).
Hai, R. et al. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat. Commun. 4, 2854 (2013).
Rogers, G. N., Pritchett, T. J., Lane, J. L. & Paulson, J. C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology 131, 394–408 (1983).
Hendricks, G. L. et al. Sialylneolacto-N-tetraose c (LSTc)-bearing liposomal decoys capture influenza A virus. J. Biol. Chem. 288, 8061–8073 (2013).
Takemoto, D. K., Skehel, J. J. & Wiley, D. C. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology 217, 452–458 (1996).
Reuter, J. D. et al. Inhibition of viral adhesion and infection by sialic-acid-conjugated dendritic polymers. Bioconjug. Chem. 10, 271–278 (1999).
Gambaryan, A. S. et al. Polymeric inhibitor of influenza virus attachment protects mice from experimental influenza infection. Antiviral Res. 55, 201–205 (2002).
Esfand, R. & Tomalia, D. A. Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications. Drug Discov. Today 6, 427–436 (2001).
Goodwin, A. P., Lam, S. S. & Fréchet, JMJ . Rapid, efficient synthesis of heterobifunctional biodegradable dendrimers. J. Am. Chem. Soc. 129, 6994–6995 (2007).
Mammen, M., Choi, S. K. & Whitesides, G. M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 37, 2754–2794 (1998).
Patke, S. et al. Design of monodisperse and well-defined polypeptide-based polyvalent inhibitors of anthrax toxin. Angew. Chem. Int. Ed. 53, 8037–8040 (2014).
Glick, G. D., Toogood, P. L., Wiley, D. C., Skehel, J. J. & Knowles, J. R. Ligand recognition by influenza virus: the binding of bivalent sialosides. J. Biol. Chem. 266, 23660–23669 (1991).
Chinarev, A. A. et al. in Sialobiology and Other Novel Forms of Glycosylation (eds Inoue, Y., Lee, Y. C. & Troy, F. A.) 135–143 (Gakushin, 1999).
Olson, S. T. & Shore, J. D. Binding of high affinity heparin to antithrombin III: characterization of the protein fluorescence enhancement. J. Biol. Chem. 256, 11065–11072 (1981).
Harris, A. et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl Acad. Sci. USA 103, 19123–19127 (2006).
Zhou, Z. H. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr. Opin. Struct. Biol. 18, 218–228 (2008).
Eichman, J. D., Bielinska, A. U., Kukowska-Latallo, J. F. & Baker, J. R. Jr The use of PAMAM dendrimers in the efficient transfer of genetic material into cells. Pharm. Sci. Technol. Today 3, 232–245 (2000).
Sakai, T. et al. Dual wavelength imaging allows analysis of membrane fusion of influenza virus inside cells. J. Virol. 80, 2013–2018 (2006).
Kwon, D., Shin, K., Kim, S. J., Lee, J. Y. & Kang, C. Mammalian pathogenesis of oseltamivir-resistant pandemic (H1N1) 2009 influenza virus isolated in South Korea. Virus Res. 185, 41–46 (2014).
Matrosovich, M. & Klenk, H. D. Natural and synthetic sialic acid-containing inhibitors of influenza virus receptor binding. Rev. Med. Virol. 13, 85–97 (2003).
Polizzotti, B. D., Maheshwari, R., Vinkenborg, J. & Kiick, K. L. Effects of saccharide spacing and chain extension on toxin inhibition by glycopolypeptides of well-defined architecture. Macromolecules 40, 7103–7110 (2007).
Choi, S. K., Mammen, M. & Whitesides, G. M. Monomeric inhibitors of influenza neuraminidase enhance the hemagglutination inhibition activities of polyacrylamides presenting multiple C-sialoside groups. Chem. Biol. 3, 97–104 (1996).
Sauter, N. K. et al. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry 31, 9609–9621 (1992).
Rowe, T. et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37, 937–943 (1999).
Isaacs, C. E. et al. Epigallocatechin gallate inactivates clinical isolates of herpes simplex virus. Antimicrob. Agents Chemother. 52, 962–970 (2008).
Jung, H. S., Komatsu, S., Ikebe, M. & Craig, R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell. 19, 3234–3242 (2008).
Acknowledgements
This work was supported by the National Research Foundation of Korea grant funded by the Korea government (MSIP) (NRF-2013R1A2A2A01068858), Marine Biotechnology Program (PJT200620) funded by Ministry of Oceans and Fisheries, and Basic Science Research Program funded by the Ministry of Education (2015R1D1A1A01060512).
Author information
Authors and Affiliations
Contributions
S.-J.K, D.H.N., and J.H.K. performed all of the experiments and analysed all data. M.D. performed the microneutralization assays. F.Z. performed the SPR analyses. E.J.P. synthesized the 6SL–PAMAM dendrimer conjugates. J.-H.P., H.Y. and C.-S.S. conducted the murine experiments. R.S.K. and J.S.D. provided critical feedback on the manuscript. S.-J.K and K.B.L. planned the experiments, interpreted the results, and wrote the manuscript with R.J.L. and J.S.D.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1970 kb)
Rights and permissions
About this article
Cite this article
Kwon, SJ., Na, D., Kwak, J. et al. Nanostructured glycan architecture is important in the inhibition of influenza A virus infection. Nature Nanotech 12, 48–54 (2017). https://doi.org/10.1038/nnano.2016.181
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2016.181
This article is cited by
-
Role of milk carbohydrates in intestinal health of nursery pigs: a review
Journal of Animal Science and Biotechnology (2022)
-
Inhibitory effect and mechanism of gelatin stabilized ferrous sulfide nanoparticles on porcine reproductive and respiratory syndrome virus
Journal of Nanobiotechnology (2022)
-
Designer DNA nanostructures for viral inhibition
Nature Protocols (2022)
-
Potent sialic acid inhibitors that target influenza A virus hemagglutinin
Scientific Reports (2021)
-
Dendrimer end-terminal motif-dependent evasion of human complement and complement activation through IgM hitchhiking
Nature Communications (2021)