Abstract
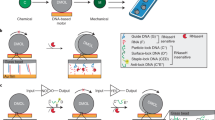
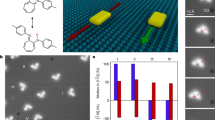
DNA-based machines that walk by converting chemical energy into controlled motion could be of use in applications such as next-generation sensors, drug-delivery platforms and biological computing. Despite their exquisite programmability, DNA-based walkers are challenging to work with because of their low fidelity and slow rates (∼1 nm min–1). Here we report DNA-based machines that roll rather than walk, and consequently have a maximum speed and processivity that is three orders of magnitude greater than the maximum for conventional DNA motors. The motors are made from DNA-coated spherical particles that hybridize to a surface modified with complementary RNA; the motion is achieved through the addition of RNase H, which selectively hydrolyses the hybridized RNA. The spherical motors can move in a self-avoiding manner, and anisotropic particles, such as dimerized or rod-shaped particles, can travel linearly without a track or external force. We also show that the motors can be used to detect single nucleotide polymorphism by measuring particle displacement using a smartphone camera.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
10 December 2015
In this Article, the wrong version of Fig. 1a was used. This error has now been corrected in all versions of the Article.
References
He, Y. & Liu, D. R. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nature Nanotech. 5, 778–782 (2010).
Lewandowski, B. et al. Sequence-specific peptide synthesis by an artificial small-molecule machine. Science 339, 189–193 (2013).
von Delius, M., Geertsema, E. M. & Leigh, D. A. A synthetic small molecule that can walk down a track. Nature Chem. 2, 96–101 (2010).
Paxton, W. F., Sundararajan, S., Mallouk, T. E. & Sen, A. Chemical locomotion. Angew. Chem. Int. Ed. 45, 5420–5429 (2006).
Pavlick, R. A., Sengupta, S., McFadden, T., Zhang, H. & Sen, A. A polymerization-powered motor. Angew. Chem. Int. Ed. 50, 9374–9377 (2011).
Orozco, J. et al. Artificial enzyme-powered microfish for water-quality testing. ACS Nano 7, 818–824 (2013).
Gu, H., Chao, J., Xiao, S. J. & Seeman, N. C. A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2010).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Wickham, S. F. et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nature Nanotech. 6, 166–169 (2011).
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA Brownian motor with coordinated legs. Science 324, 67–71 (2009).
Wickham, S. F. et al. A DNA-based molecular motor that can navigate a network of tracks. Nature Nanotech. 7, 169–173 (2012).
Cha, T. G. et al. A synthetic DNA motor that transports nanoparticles along carbon nanotubes. Nature Nanotech. 9, 39–43 (2014).
Yin, P., Choi, H. M. T., Calvert, C. R. & Pierce, N. A. Programming biomolecular self-assembly pathways. Nature 451, 318–U314 (2008).
Bath, J., Green, S. J. & Turberfield, A. J. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. 44, 4358–4361 (2005).
Pei, R. et al. Behavior of polycatalytic assemblies in a substrate-displaying matrix. J. Am. Chem. Soc. 128, 12693–12699 (2006).
Perl, A. et al. Gradient-driven motion of multivalent ligand molecules along a surface functionalized with multiple receptors. Nature Chem. 3, 317–322 (2011).
Fang, S., Lee, H. J., Wark, A. W., Kim, H. M. & Corn, R. M. Determination of ribonuclease H surface enzyme kinetics by surface plasmon resonance imaging and surface plasmon fluorescence spectroscopy. Anal. Chem. 77, 6528–6534 (2005).
Yehl, K. et al. Catalytic deoxyribozyme-modified nanoparticles for RNAi-independent gene regulation. ACS Nano 6, 9150–9157 (2012).
Liu, Y., Yehl, K., Narui, Y. & Salaita, K. Tension sensing nanoparticles for mechano-imaging at the living/nonliving interface. J. Am. Chem. Soc. 135, 5320–5323 (2013).
Liu, Y. et al. Nanoparticle tension probes patterned at the nanoscale: impact of integrin clustering on force transmission. Nano Lett. 14, 5539–5546 (2014).
Sbalzarini, I. F. & Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151, 182–195 (2005).
Gal, N., Lechtman-Goldstein, D. & Weihs, D. Particle tracking in living cells: a review of the mean square displacement method and beyond. Rheol. Acta 52, 425–443 (2013).
Domb, C., Gillis, J. & Wilmers, G. On shape and configuration of polymer molecules. Proc. Phys. Soc. 85, 625 (1965).
Amit, D. J., Parisi, G. & Peliti, L. Asymptotic-behavior of the true self-avoiding walk. Phys. Rev. B 27, 1635–1645 (1983).
Obukhov, S. P. & Peliti, L. Renormalization of the true self-avoiding walk. J. Phys. A 16, L147–L151 (1983).
Family, F. & Daoud, M. Experimental realization of true self-avoiding walks. Phys. Rev. B 29, 1506–1507 (1984).
Diehl, M. R., Zhang, K., Lee, H. J. & Tirrell, D. A. Engineering cooperativity in biomotor–protein assemblies. Science 311, 1468–1471 (2006).
Østergaard, M. E. et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 41, 9634–9650 (2013).
Berna, J. et al. Macroscopic transport by synthetic molecular machines. Nature Mater. 4, 704–710 (2005).
Eelkema, R. et al. Nanomotor rotates microscale objects. Nature 440, 163–163 (2006).
Liu, Y. et al. Linear artificial molecular muscles. J. Am. Chem. Soc. 127, 9745–9759 (2005).
Wang, W., Chiang, T. Y., Velegol, D. & Mallouk, T. E. Understanding the efficiency of autonomous nano- and microscale motors. J. Am. Chem. Soc. 135, 10557–10565 (2013).
Peterson, A. W., Heaton, R. J. & Georgiadis, R. M. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 29, 5163–5168 (2001).
Yan, L., Zhao, X. M. & Whitesides, G. M. Patterning a preformed, reactive SAM using microcontact printing. J. Am. Chem. Soc. 120, 6179–6180 (1998).
Zhang, Y., Ge, C. H., Zhu, C. & Salaita, K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nature Commun. 5, 5167 (2014).
Johnson, K. A. & Goody, R. S. The original Michaelis constant: translation of the 1913 Michaelis–Menten paper. Biochemistry 50, 8264–8269 (2011).
Acknowledgements
K.S. is grateful for support from the National Institutes of Health through R01-GM097399, the Alfred P. Sloan Research Fellowship, the Camille–Dreyfus Teacher–Scholar Award and the National Science Foundation (NSF) CAREER Award (1350829). K.Y. thanks the ARCS Foundation for their support and V. Pui-Yan Ma for generating Fig. 1. We also thank S. Urazhdin for access to the thermal evaporator and M. Grover and D. Stabley for helpful discussions. E.R.W. was funded by the NSF (CMMI-1250235) and S.V. was funded by Emory University. This research project was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core.
Author information
Authors and Affiliations
Contributions
K.Y. conducted all the experiments and analysis, A.M. performed the simulations and theoretical validation, S.V. helped in the data analysis and validation of the theoretical model, Y.Z. helped with particle functionalization, Y.L. collected SIM data, M.F. assisted with SNP detection, K.S. and K.Y. wrote the manuscript with input from A.M. and E.R.W., K.S. oversaw all the aspects of the work and E.R.W. supervised and discussed the experiments with S.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Information (PDF 3610 kb)
Supplementary Movie 1
Supplementary Movie 1 (AVI 2675 kb)
Supplementary Movie 2
Supplementary Movie 2 (AVI 339 kb)
Supplementary Movie 3
Supplementary Movie 3 (AVI 250 kb)
Supplementary Movie 4
Supplementary Movie 4 (MP4 983 kb)
Supplementary Movie 5
Supplementary Movie 5 (AVI 2455 kb)
Supplementary Movie 6
Supplementary Movie 6 (AVI 310 kb)
Supplementary Movie 7
Supplementary Movie 7 (AVI 2476 kb)
Supplementary Movie 8
Supplementary Movie 8 (AVI 4785 kb)
Supplementary Movie 9
Supplementary Movie 9 (AVI 380 kb)
Supplementary Movie 10
Supplementary Movie 10 (AVI 2668 kb)
Supplementary Movie 11
Supplementary Movie 11 (AVI 4767 kb)
Supplementary Movie 12
Supplementary Movie 12 (AVI 233 kb)
Supplementary Movie 13
Supplementary Movie 13 (AVI 260 kb)
Supplementary Movie 14
Supplementary Movie 14 (AVI 0 kb)
Rights and permissions
About this article
Cite this article
Yehl, K., Mugler, A., Vivek, S. et al. High-speed DNA-based rolling motors powered by RNase H. Nature Nanotech 11, 184–190 (2016). https://doi.org/10.1038/nnano.2015.259
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2015.259
This article is cited by
-
Enzyme-driven Nanorobots Walking Along Predesigned Tracks on the DNA Origami for Cargo Transport and Catalysis
Chemical Research in Chinese Universities (2024)
-
High-throughput microbead assay system with a portable, cost-effective Wi-Fi imaging module, and disposable multi-layered microfluidic cartridges for virus and microparticle detection, and tracking
Biomedical Microdevices (2023)
-
Bispecific aptamer-initiated 3D DNA nanomotor biosensor powered by DNAzyme and entropy-driven circuit for sensitive and specificity detection of lysozyme
Nano Research (2023)
-
Sustained unidirectional rotation of a self-organized DNA rotor on a nanopore
Nature Physics (2022)
-
Functional DNA-based cytoskeletons for synthetic cells
Nature Chemistry (2022)