Abstract
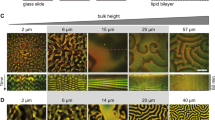
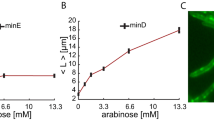
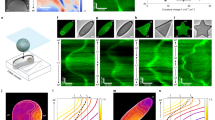
The boundary of a cell defines the shape and scale of its subcellular organization. However, the effects of the cell's spatial boundaries as well as the geometry sensing and scale adaptation of intracellular molecular networks remain largely unexplored. Here, we show that living bacterial cells can be ‘sculpted’ into defined shapes, such as squares and rectangles, which are used to explore the spatial adaptation of Min proteins that oscillate pole-to-pole in rod-shaped Escherichia coli to assist cell division. In a wide geometric parameter space, ranging from 2 × 1 × 1 to 11 × 6 × 1 μm3, Min proteins exhibit versatile oscillation patterns, sustaining rotational, longitudinal, diagonal, stripe and even transversal modes. These patterns are found to directly capture the symmetry and scale of the cell boundary, and the Min concentration gradients scale with the cell size within a characteristic length range of 3–6 μm. Numerical simulations reveal that local microscopic Turing kinetics of Min proteins can yield global symmetry selection, gradient scaling and an adaptive range, when and only when facilitated by the three-dimensional confinement of the cell boundary. These findings cannot be explained by previous geometry-sensing models based on the longest distance, membrane area or curvature, and reveal that spatial boundaries can facilitate simple molecular interactions to result in far more versatile functions than previously understood.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
25 June 2015
In the version of this Article previously published online the Methods section was inadvertently omitted. This has been corrected for all versions of the Article.
References
Turing, A. M. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72 (1952).
Gierer, A. & Meinhardt, H. A theory of biological pattern formation. Kybernetik 12, 30–39 (1972).
Meinhardt, H. & de Boer, P. A. J. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl Acad. Sci. USA 98, 14202–14207 (2001).
Huang, K. C., Meir, Y. & Wingreen, N. S. Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones. Proc. Natl Acad. Sci. USA 100, 12724–12728 (2003).
Goryachev, A. B. & Pokhilko, A. V. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 582, 1437–1443 (2008).
Kondo, S. & Miura, T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620 (2010).
Halatek, J. & Frey, E. Highly canalized MinD transfer and MinE sequestration explain the rigin of robust MinCDE-protein dynamics. Cell Rep. 1, 741–752 (2012).
Noorduin, W. L., Grinthal, A., Mahadevan, L. & Aizenberg, J. Rationally designed complex, hierarchical microarchitectures. Science 340, 832–837 (2013).
Moseley, J. B. & Nurse, P. Cell division intersects with cell geometry. Cell 142, 184–188 (2010).
Raskin, D. M. & de Boer, P. A. J. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA 96, 4971–4976 (1999).
Hu, Z. & Lutkenhaus, J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34, 82–90 (1999).
Ben-Jacob, E. Bacterial self-organization: co-enhancement of complexification and adaptability in a dynamic environment. Phil. Trans. R. Soc. Lond. A 361, 1283–1312 (2003).
Männik, J. et al. Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proc. Natl Acad. Sci. USA 109, 6957–6962 (2012).
Young, K. D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703 (2006).
Hu, Z. & Lutkenhaus, J. Topological regulation of cell division in E. coli: spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7, 1337–1343 (2001).
Lackner, L. L., Raskin, D. M. & de Boer, P. A. J. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J. Bacteriol. 185, 735–749 (2003).
Loose, M., Fischer-Friedrich, E., Ries, J., Kruse, K. & Schwille, P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792 (2008).
Ivanov, V. & Mizuuchi, K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc. Natl Acad. Sci. USA 107, 8071–8078 (2010).
Schweizer, J. et al. Geometry sensing by self-organized protein patterns. Proc. Natl Acad. Sci. USA 109, 15283–15288 (2012).
Zieske, K. & Schwille, P. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife 3, e03949 (2014).
Vecchiarelli, A. G., Li, M., Mizuuchi, M. & Mizuuchi, K. Differential affinities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol. Microbiol. 93, 453–463 (2014).
Corbin, B. D., Yu, X-C. & Margolin, W. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 21, 1998–2008 (2002).
Varma, A., Huang, K. C. & Young, K. D. The Min system as a general cell geometry detection mechanism: branch lengths in Y-shaped Escherichia coli cells affect Min oscillation patterns and division dynamics. J. Bacteriol. 190, 2106–2117 (2008).
Renner, L. D. & Weibel, D. B. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl Acad. Sci. USA 108, 6264–6269 (2011).
Typas, A., Banzhaf, M., Gross, C. A. & Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nature Rev. Microbiol. 10, 123–136 (2012).
Takeuchi, S., DiLuzio, W. R., Weibel, D. B. & Whitesides, G. M. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 5, 1819–1823 (2005).
Männik, J., Driessen, R., Galajda, P., Keymer, J. E. & Dekker, C. Bacterial growth and motility in sub-micron constrictions. Proc. Natl Acad. Sci. USA 106, 14861–14866 (2009).
Cabeen, M. T. et al. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28, 1208–1219 (2009).
Fischer-Friedrich, E., Meacci, G., Lutkenhaus, J., Chaté, H. & Kruse, K. Intra- and intercellular fluctuations in Min-protein dynamics decrease with cell length. Proc. Natl Acad. Sci. USA 107, 6134–6139 (2010).
Shannon, C. E. A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948).
Yu, X-C. & Margolin, W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32, 315–326 (1999).
Fange, D. & Elf, J. Noise-induced Min phenotypes in E. coli. PLoS Comput. Biol. 2, e80 (2006).
Ben-Zvi, D., Shilo, B-Z., Fainsod, A. & Barkai, N. Scaling of the BMP activation gradient in Xenopus embryos. Nature 453, 1205–1211 (2008).
Ben-Zvi, D. & Barkai, N. Scaling of morphogen gradients by an expansion-repression integral feedback control. Proc. Natl Acad. Sci. USA 107, 6924–6929 (2010).
Lauschke, V. M., Tsiairis, C. D., Francois, P. & Aulehla, A. Scaling of embryonic patterning based on phase-gradient encoding. Nature 493, 101–105 (2013).
Averbukh, I., Ben-Zvi, D., Mishra, S. & Barkai, N. Scaling morphogen gradients during tissue growth by a cell division rule. Development 141, 2150–2156 (2014).
Loose, M., Fischer-Friedrich, E., Herold, C., Kruse, K. & Schwille, P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nature Struct. Mol. Biol. 18, 577–583 (2011).
Hoffmann, M. & Schwarz, U. S. Oscillations of Min-proteins in micropatterned environments: a three-dimensional particle-based stochastic simulation approach. Soft Matter 10, 2388–2396 (2014).
Hsieh, C-W. et al. Direct MinE–membrane interaction contributes to the proper localization of MinDE in E. coli. Mol. Microbiol. 75, 499–512 (2010).
Park, K-T. et al. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell 146, 396–407 (2011).
Leisch, N. et al. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr. Biol. 22, R831–R832 (2012).
Pende, N. et al. Size-independent symmetric division in extraordinarily long cells. Nature Commun. 5, 4803 (2014).
Vicker, M. G. F-actin assembly in Dictyostelium cell locomotion and shape oscillations propagates as a self-organized reaction-diffusion wave. FEBS Lett. 510, 5–9 (2002).
Chau, A. H., Walter, J. M., Gerardin, J., Tang, C. & Lim, W. A. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell 151, 320–332 (2012).
Shapiro, L., McAdams, H. H. & Losick, R. Why and how bacteria localize proteins. Science 326, 1225–1228 (2009).
Huang, Z., Pedaci, F., van Oene, M., Wiggin, M. J. & Dekker, N. H. Electron beam fabrication of birefringent microcylinders. ACS Nano 5, 1418–1427 (2011).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Wu, F., van Rijn, E., van Schie, B. G. C., Keymer, J. E. & Dekker, C. Multicolor imaging of bacterial nucleoid and division proteins with blue, orange and near-infrared fluorescent proteins. Front. Microbiol. 6, 607 (2015).
Loew, L. M. & Schaff, J. C. The virtual cell: a software environment for computational cell biology. Trends Biotechnol. 19, 401–406 (2001).
Acknowledgements
The authors thank E. van Rijn, D. de Graaff, W. Postek, J. van der Does, J. Kerssenmakers and Z. Huang for technical assistance, Y. Caspi, Y-L. Shih, A. Lindert, L. Rothfield, A. Meyer and C. Plesa for materials, E. Frey and J. Halatek for discussions on their model, C. Danelon and F. Hol for discussions, and the CSHL Computational Cell Biology Summer School and VirtualCell (NIH grant P41-GM103313). This work was partly supported by the Netherlands Organisation for Scientific Research (NWO/OCW) as part of the Frontiers of Nanoscience programme, NanoNextNL programme 3B (F.W.) and European Research Council NanoforBio no. 247072 (C.D.).
Author information
Authors and Affiliations
Contributions
F.W., J.E.K. and C.D. conceived the experiments, discussed the work and wrote the paper. F.W. and B.v.S performed the experiments. F.W. analysed the data, derived the mechanisms and carried out computer simulations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Information (PDF 8971 kb)
Supplementary Movie 1
Supplementary Movie 1 (AVI 131 kb)
Supplementary Movie 2
Supplementary Movie 2 (AVI 1605 kb)
Supplementary Movie 3
Supplementary Movie 3 (AVI 425 kb)
Supplementary Movie 4
Supplementary Movie 4 (AVI 1759 kb)
Supplementary Movie 5
Supplementary Movie 5 (AVI 2293 kb)
Supplementary Movie 6
Supplementary Movie 6 (AVI 6953 kb)
Supplementary Movie 7
Supplementary Movie 7 (AVI 1585 kb)
Supplementary Movie 8
Supplementary Movie 8 (AVI 635 kb)
Supplementary Movie 9
Supplementary Movie 9 (AVI 1699 kb)
Supplementary Movie 10
Supplementary Movie 10 (AVI 2296 kb)
Supplementary Movie 11
Supplementary Movie 11 (AVI 131 kb)
Supplementary Movie 12
Supplementary Movie 12 (AVI 1177 kb)
Rights and permissions
About this article
Cite this article
Wu, F., van Schie, B., Keymer, J. et al. Symmetry and scale orient Min protein patterns in shaped bacterial sculptures. Nature Nanotech 10, 719–726 (2015). https://doi.org/10.1038/nnano.2015.126
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2015.126
This article is cited by
-
Bacterial cell-size changes resulting from altering the relative expression of Min proteins
Nature Communications (2023)
-
Asymmetric peptidoglycan editing generates cell curvature in Bdellovibrio predatory bacteria
Nature Communications (2022)
-
Artificial modulation of cell width significantly affects the division time of Escherichia coli
Scientific Reports (2020)
-
Synthetic cell division via membrane-transforming molecular assemblies
BMC Biology (2019)
-
Direct imaging of the circular chromosome in a live bacterium
Nature Communications (2019)