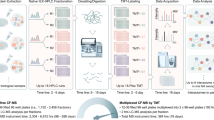
Abstract
Interactomes are often measured using affinity purification–mass spectrometry (AP-MS) or yeast two-hybrid approaches, but these methods do not provide stoichiometric or temporal information. We combine quantitative proteomics and size-exclusion chromatography to map 291 coeluting complexes. This method allows mapping of an interactome to the same depth and accuracy as AP-MS with less work and without overexpression or tagging. The use of triplex labeling enables monitoring of interactome rearrangements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Blume-Jensen, P. & Hunter, T. Nature 411, 355–365 (2001).
Gavin, A.-C. et al. Nature 440, 631–636 (2006).
Uetz, P. et al. Nature 403, 623–627 (2000).
Werner, J.N. et al. Proc. Natl. Acad. Sci. USA 106, 7858–7863 (2009).
Andersen, J.S. et al. Nature 426, 570–574 (2003).
Foster, L.J. et al. Cell 125, 187–199 (2006).
Olinares, P.D., Ponnala, L. & van Wijk, K.J. Mol. Cell. Proteomics 9, 1594–1615 (2010).
Wang, X. & Huang, L. Mol. Cell. Proteomics 7, 46–57 (2008).
Jin, J. et al. Curr. Biol. 14, 1436–1450 (2004).
Tai, H.-C., Besche, H., Goldberg, A.L. & Schuman, E.M. Front. Mol. Neurosci. 3 (2010); doi:10.3389/fnmol.2010.00012.
Vuori, K. & Ruoslahti, E. Science 266, 1576–1578 (1994).
Bache, K.G., Raiborg, C., Mehlum, A. & Stenmark, H. J. Biol. Chem. 278, 12513–12521 (2003).
Olsen, J.V. et al. Cell 127, 635–648 (2006).
Blagoev, B., Ong, S.-E., Kratchmarova, I. & Mann, M. Nat. Biotechnol. 22, 1139–1145 (2004).
Argenzio, E. et al. Mol. Syst. Biol. 7, 462 (2011).
Ewing, R.M. et al. Mol. Syst. Biol. 3, 89 (2007).
Sancak, Y. et al. Mol. Cell 25, 903–915 (2007).
Rogers, L.D. & Foster, L.J. Proc. Natl. Acad. Sci. USA 104, 18520–18525 (2007).
Rappsilber, J., Ishihama, Y. & Mann, M. Anal. Chem. 75, 663–670 (2003).
Cox, J. & Mann, M. Nat. Biotechnol. 26, 1367–1372 (2008).
Ruepp, A. et al. Nucleic Acids Res. 38, D497–D501 (2010).
Davis, J. & Goadrich, M. in Proc. 23rd Int. Conf. Mach. Learn. 233–240 (ACM, 2006).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J.V. & Mann, M. Nat. Protoc. 1, 2856–2860 (2006).
Aranda, B. et al. Nucleic Acids Res. 38, D525–D531 (2010).
Acknowledgements
The authors thank members of the Foster group for discussions and advice. This work was supported by a grant from the Canadian Institutes for Health Research to L.J.F. (MOP-77688). L.J.F. is supported by the Canada Research Chairs program and A.R.K. is supported by the Danish Agency for Science Technology and Innovation. Mass spectrometry infrastructure used in this work was supported by the Canada Foundation for Innovation, British Columbia Knowledge Development Fund and BC Proteomics Network.
Author information
Authors and Affiliations
Contributions
A.R.K. conceived of and performed the experiments; A.R.K., J.G. and L.J.F. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 837 kb)
Supplementary Table 1a
Raw chromatograms for proteins identified in the three biological replicates (XLS 2363 kb)
Supplementary Table 2
Limits applied for the three different biological replicates (XLS 19 kb)
Supplementary Table 3
Binary interactions identified (XLS 2762 kb)
Supplementary Table 4
Protein complexes identified (XLS 206 kb)
Supplementary Table 5
Proteins that did change elution time following addition of antibodies against 14-3-3A (XLS 19 kb)
Supplementary Table 6
Proteins identified to have temporal changes after EGF stimulation (XLS 45 kb)
Supplementary Table 7
Proteins that changed elution time following addition of antibody against HRS (XLS 19 kb)
Rights and permissions
About this article
Cite this article
Kristensen, A., Gsponer, J. & Foster, L. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods 9, 907–909 (2012). https://doi.org/10.1038/nmeth.2131
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2131
This article is cited by
-
DIP-MS: ultra-deep interaction proteomics for the deconvolution of protein complexes
Nature Methods (2024)
-
Biological big-data sources, problems of storage, computational issues, and applications: a comprehensive review
Knowledge and Information Systems (2024)
-
Molecular complex detection in protein interaction networks through reinforcement learning
BMC Bioinformatics (2023)
-
Co-fractionation–mass spectrometry to characterize native mitochondrial protein assemblies in mammalian neurons and brain
Nature Protocols (2023)
-
Mapping protein states and interactions across the tree of life with co-fractionation mass spectrometry
Nature Communications (2023)