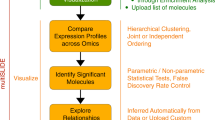
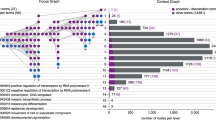
Abstract
Cytoscape is open-source software for integration, visualization and analysis of biological networks. It can be extended through Cytoscape plugins, enabling a broad community of scientists to contribute useful features. This growth has occurred organically through the independent efforts of diverse authors, yielding a powerful but heterogeneous set of tools. We present a travel guide to the world of plugins, covering the 152 publicly available plugins for Cytoscape 2.5–2.8. We also describe ongoing efforts to distribute, organize and maintain the quality of the collection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cline, M.S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Yeung, N., Cline, M.S., Kuchinsky, A., Smoot, M.E. & Bader, G.D. Exploring biological networks with Cytoscape software. Curr. Protoc. Bioinformatics 23, 8.13 (2008).
Hermjakob, H. et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 22, 177–183 (2004).
Demir, E. et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 28, 935–942 (2010).
Hucka, M. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531 (2003).
Stark, C. et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 39, D698–D704 (2011).
Gao, J. et al. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics 25, 137–138 (2009).
Pentchev, K., Ono, K., Herwig, R., Ideker, T. & Kamburov, A. Evidence mining and novelty assessment of protein-protein interactions with the ConsensusPathDB plugin for Cytoscape. Bioinformatics 26, 2796–2797 (2010).
Hernandez-Toro, J., Prieto, C. & De las Rivas, J. APID2NET: unified interactome graphic analyzer. Bioinformatics 23, 2495–2497 (2007).
Aranda, B. et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods 8, 528–529 (2011).
Gao, J. et al. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics 26, 971–973 (2010).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Ma, H. et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 3, 135 (2007).
Pico, A.R. et al. WikiPathways: pathway editing for the people. PLoS Biol. 6, e184 (2008).
Cerami, E.G. et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 39, D685–D690 (2011).
Vailaya, A. et al. An architecture for biological information extraction and representation. Bioinformatics 21, 430–438 (2005).
Hamosh, A., Scott, A.F., Amberger, J.S., Bocchini, C.A. & McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 (2005).
Cusick, M.E. et al. Literature-curated protein interaction datasets. Nat. Methods 6, 39–46 (2009).
Montojo, J. et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928 (2010).
Lee, P.H. & Lee, D. Modularized learning of genetic interaction networks from biological annotations and mRNA expression data. Bioinformatics 21, 2739–2747 (2005).
Henry, C.S. et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982 (2010).
Assenov, Y., Ramirez, F., Schelhorn, S.E., Lengauer, T. & Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 24, 282–284 (2008).
Jeong, H., Mason, S.P., Barabasi, A.L. & Oltvai, Z.N. Lethality and centrality in protein networks. Nature 411, 41–42 (2001).
Ladha, J. et al. Glioblastoma-specific protein interaction network identifies PP1A and CSK21 as connecting molecules between cell cycle-associated genes. Cancer Res. 70, 6437–6447 (2010).
Scardoni, G., Petterlini, M. & Laudanna, C. Analyzing biological network parameters with CentiScaPe. Bioinformatics 25, 2857–2859 (2009).
Enright, A.J., Van Dongen, S. & Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Frey, B.J. & Dueck, D. Clustering by passing messages between data points. Science 315, 972–976 (2007).
Bader, G.D. & Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2 (2003).
Rivera, C.G., Vakil, R. & Bader, J.S. NeMo: Network Module identification in Cytoscape. BMC Bioinformatics 11 (suppl. 1), S61 (2010).
Rhrissorrakrai, K. & Gunsalus, K.C. MINE: Module Identification in Networks. BMC Bioinformatics 12, 192 (2011).
Morris, J.H. et al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12, 436 (2011).
Li, X., Wu, M., Kwoh, C.K. & Ng, S.K. Computational approaches for detecting protein complexes from protein interaction networks: a survey. BMC Genomics 11 (suppl. 1), S3 (2010).
Moschopoulos, C.N. et al. Which clustering algorithm is better for predicting protein complexes? BMC Res. Notes 4, 549 (2011).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Smoot, M., Ono, K., Ideker, T. & Maere, S. PiNGO: a Cytoscape plugin to find candidate genes in biological networks. Bioinformatics 27, 1030–1031 (2011).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5, e13984 (2010).
Oesper, L., Merico, D., Isserlin, R. & Bader, G.D. WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol. Med. 6, 7 (2011).
Haider, S. et al. BioMart Central Portal—unified access to biological data. Nucleic Acids Res. 37, W23–W27 (2009).
van Iersel, M.P. et al. The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services. BMC Bioinformatics 11, 5 (2010).
Kincaid, R., Kuchinsky, A. & Creech, M. VistaClara: an expression browser plug-in for Cytoscape. Bioinformatics 24, 2112–2114 (2008).
Yang, L., Walker, J.R., Hogenesch, J.B. & Thomas, R.S. NetAtlas: a Cytoscape plugin to examine signaling networks based on tissue gene expression. In Silico Biol. 8, 47–52 (2008).
Xia, T., Hemert, J.V. & Dickerson, J.A. OmicsAnalyzer: a Cytoscape plug-in suite for modeling omics data. Bioinformatics 26, 2995–2996 (2010).
Ideker, T., Ozier, O., Schwikowski, B. & Siegel, A.F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 18 (suppl. 1), S233–S240 (2002).
Alcaraz, N., Kücük, H., Weile, J., Wipat, A. & Baumbach, J. KeyPathwayMiner: detecting case-specific biological pathways using expression data. Internet Math. 7, 299–313 (2011).
Chuang, H.Y., Lee, E., Liu, Y.T., Lee, D. & Ideker, T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 3, 140 (2007).
Guziolowski, C., Bourde, A., Moreews, F. & Siegel, A. BioQuali Cytoscape plugin: analysing the global consistency of regulatory networks. BMC Genomics 10, 244 (2009).
Warsow, G. et al. ExprEssence—revealing the essence of differential experimental data in the context of an interaction/regulation net-work. BMC Syst. Biol. 4, 164 (2010).
Li, F. et al. PerturbationAnalyzer: a tool for investigating the effects of concentration perturbation on protein interaction networks. Bioinformatics 26, 275–277 (2010).
Emig, D. et al. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 38, W755–W762 (2010).
Wang, L., Khankhanian, P., Baranzini, S.E. & Mousavi, P. iCTNet: a Cytoscape plugin to produce and analyze integrative complex traits networks. BMC Bioinformatics 12, 380 (2011).
Singhal, M. & Domico, K. CABIN: collective analysis of biological interaction networks. Comput. Biol. Chem. 31, 222–225 (2007).
Petyuk, V.A. et al. Characterization of the mouse pancreatic islet proteome and comparative analysis with other mouse tissues. J. Proteome Res. 7, 3114–3126 (2008).
Woźniak, M., Tiuryn, J. & Dutkowski, J. MODEVO: exploring modularity and evolution of protein interaction networks. Bioinformatics 26, 1790–1791 (2010).
Hao, Y. et al. OrthoNets: simultaneous visual analysis of orthologs and their interaction neighborhoods across different organisms. Bioinformatics 27, 883–884 (2011).
Srivas, R. et al. Assembling global maps of cellular function through integrative analysis of physical and genetic networks. Nat. Protoc. 6, 1308–1323 (2011).
Shannon, P.T., Reiss, D.J., Bonneau, R. & Baliga, N.S. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics 7, 176 (2006).
Wittkop, T. et al. Comprehensive cluster analysis with Transitivity Clustering. Nat. Protoc. 6, 285–295 (2011).
Morris, J.H., Huang, C.C., Babbitt, P.C. & Ferrin, T.E. structureViz: linking Cytoscape and UCSF Chimera. Bioinformatics 23, 2345–2347 (2007).
Doncheva, N.T., Klein, K., Domingues, F.S. & Albrecht, M. Analyzing and visualizing residue networks of protein structures. Trends Biochem. Sci. 36, 179–182 (2011).
Erhard, F., Friedel, C.C. & Zimmer, R. FERN – a Java framework for stochastic simulation and evaluation of reaction networks. BMC Bioinformatics 9, 356 (2008).
Merico, D., Gfeller, D. & Bader, G.D. How to visually interpret biological data using networks. Nat. Biotechnol. 27, 921–924 (2009).
Acknowledgements
Work on this review was funded by the National Resource for Network Biology (P41 GM103504) and the San Diego Center for Systems Biology (P50 GM085764). We thank J. Dutkowski, D. Emig and G. Hannum for advice and critical reading of the manuscript. Finally, the greatest thanks go to all of the plugin developers who have enriched the Cytoscape user experience with their ideas. We apologize to those plugin authors whose excellent work was not covered here because of space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 888 kb)
Supplementary Data
Comprehensive list of Cytoscape plugins that we reviewed and tagged. (XLS 56 kb)
Rights and permissions
About this article
Cite this article
Saito, R., Smoot, M., Ono, K. et al. A travel guide to Cytoscape plugins. Nat Methods 9, 1069–1076 (2012). https://doi.org/10.1038/nmeth.2212
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2212
This article is cited by
-
Comprehensive analysis of miRNA profiling in Schistosoma mekongi across life cycle stages
Scientific Reports (2024)
-
Regulation of coconut somatic embryogenesis: decoding the role of long non-coding RNAs
Plant Biotechnology Reports (2024)
-
Magnetic transferrin nanoparticles (MTNs) assay as a novel isolation approach for exosomal biomarkers in neurological diseases
Biomaterials Research (2023)
-
Identification of key genes and the pathophysiology associated with allergen-specific immunotherapy for allergic rhinitis
BMC Immunology (2023)
-
Reduced LHFPL3-AS2 lncRNA expression is linked to altered epithelial polarity and proliferation, and to ileal ulceration in Crohn disease
Scientific Reports (2023)