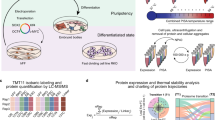
Abstract
Combining high-mass-accuracy mass spectrometry, isobaric tagging and software for multiplexed, large-scale protein quantification, we report deep proteomic coverage of four human embryonic stem cell and four induced pluripotent stem cell lines in biological triplicate. This 24-sample comparison resulted in a very large set of identified proteins and phosphorylation sites in pluripotent cells. The statistical analysis afforded by our approach revealed subtle but reproducible differences in protein expression and protein phosphorylation between embryonic stem cells and induced pluripotent cells. Merging these results with RNA-seq analysis data, we found functionally related differences across each tier of regulation. We also introduce the Stem Cell–Omics Repository (SCOR), a resource to collate and display quantitative information across multiple planes of measurement, including mRNA, protein and post-translational modifications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yu, J. et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 (2009).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Chin, M.H. et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5, 111–123 (2009).
Guenther, M.G. et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7, 249–257 (2010).
Chin, M.H., Pellegrini, M., Plath, K. & Lowry, W.E. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell 7, 263–269 (2010).
Bock, C. et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439–452 (2011).
Stadtfeld, M. et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181 (2010).
Doi, A. et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41, 1350–1353 (2009).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Olsen, J.V. et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 (2007).
McAlister, G.C., Phanstiel, D., Wenger, C.D., Lee, M.V. & Coon, J.J. Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification. Anal. Chem. 82, 316–322 (2010).
Olsen, J.V. et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 (2009).
Nagaraj, N., D′Souza, R.C.J., Cox, J., Olsen, J.V. & Mann, M. Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9, 6786–6794 (2010).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003).
Ross, P.L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 (2004).
Choe, L. et al. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics 7, 3651–3660 (2007).
Ow, S.Y. et al. iTRAQ underestimation in simple and complex mixtures: the good, the bad and the ugly. J. Proteome Res. 8, 5347–5355 (2009).
Wenger, C.D., Phanstiel, D.H., Lee, M.V., Bailey, D.J. & Coon, J.J. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 6, 1064–1074 (2011).
Shadforth, I.P., Dunkley, T.P.J., Lilley, K.S. & Bessant, C. i-Tracker: for quantitative proteomics using iTRAQ (TM). BMC Genomics 6, 145 (2005).
Griffin, T.J. et al. iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J. Proteome Res. 6, 4200–4209 (2007).
Becker, K.A., Stein, J.L., Lian, J.B., van Wijnen, A.J. & Stein, G.S. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J. Cell. Physiol. 210, 517–526 (2007).
Xue, Y. et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7, 1598–1608 (2008).
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57, 289–300 (1995).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Frye, M. & Watt, F.M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 16, 971–981 (2006).
Singh, M.K. et al. The T-box transcription factor Tbx15 is required for skeletal development. Mech. Dev. 122, 131–144 (2005).
Dong, F. et al. Pitx2 promotes development of splanchnic mesoderm-derived branchiomeric muscle. Development 133, 4891–4899 (2006).
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 (2010).
Polo, J.M. et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855 (2010).
Hu, B.Y. et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 107, 4335–4340 (2010).
Siu, I.M. et al. Coexpression of neuronatin splice forms promotes medulloblastoma growth. Neuro-oncol. 10, 716–724 (2008).
Hargrave, M. et al. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev. Dyn. 210, 79–86 (1997).
Kawano, Y. et al. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol. Cell. Biol. 25, 9920–9935 (2005).
Zhu, H., Coppinger, J.A., Jang, C.Y., Yates, J.R. III & Fang, G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol. 183, 835–848 (2008).
Bourke, E., Brown, J.A.L., Takeda, S., Hochegger, H. & Morrison, C.G. DNA damage induces Chk1-dependent threonine-160 phosphorylation and activation of Cdk2. Oncogene 29, 616–624 (2010).
Ludwig, T.E. et al. Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 24, 185–187 (2006).
Sengupta, S. et al. Highly consistent, fully representative mRNA-Seq libraries from ten nanograms of total RNA. Biotechniques 49, 898–904 (2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10 R25 (2009).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Good, D.M. et al. Post-acquisition ETD spectral processing for increased peptide identifications. J. Am. Soc. Mass Spectrom. 20, 1435–1440 (2009).
Geer, L.Y. et al. Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 (2004).
Kersey, P.J. et al. The International Protein Index: An integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004).
Nesvizhskii, A.I. & Aebersold, R. Interpretation of shotgun proteomic data - The protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 (2005).
Swaney, D.L., Wenger, C.D., Thomson, J.A. & Coon, J.J. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 106, 995–1000 (2009).
Acknowledgements
We acknowledge A.J. Bureta for helping with and providing illustrations, A. Williams and K. Eastman for editing the text, G.McAlister for assistance with instrumentation, J. Yu for providing transcriptomic data before publication and Thomson lab members for critical reading and discussion of this manuscript. This work was supported by the University of Wisconsin, the Beckman Foundation and US National Institutes of Health (NIH) grants R01GM080148 (to J.J.C.) and P01GM081629 (to J.A.T. and J.J.C.). D.H.P. acknowledges support from a NIH predoctoral traineeship, the Genomic Sciences Training Program, NIH grant 5T32HG002760.
Author information
Authors and Affiliations
Contributions
D.H.P. designed research, prepared samples, performed mass spectrometry, wrote software, analyzed data and wrote the manuscript. J.B. designed research, grew cells, prepared samples, analyzed data and wrote the manuscript. C.D.W. wrote software. S.T. and V.R. analyzed data. M.D.P. grew cells. D.J.B. designed websites. D.L.S. helped with phosphorylation analysis. M.A.T. optimized the labeling procedure. J.M.B. performed RNA sequencing. R.S. designed research and analyzed data. J.A.T. and J.J.C. designed research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.A.T. is a founder, stockowner, consultant and board member of Cellular Dynamics International (CDI), and serves as scientific advisor to and has financial interests in Tactics II Stem Cell Ventures. J.J.C. is a consultant for Thermo Fisher Scientific.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 4,8,9 (PDF 9330 kb)
Supplementary Table 1
Proteomic identification and quantification. (XLSX 5966 kb)
Supplementary Table 2
Phosphoproteomic identification and quantification. (XLSX 9623 kb)
Supplementary Table 3
Enrichment analysis from fourplex experiment. (XLSX 696 kb)
Supplementary Table 5
Transcriptomic identification and quantification. (XLSX 5051 kb)
Supplementary Table 6
Transcripts, proteins, and phosphorylation sites that differ between ESCs and iPSCs. (XLSX 1265 kb)
Supplementary Table 7
Enrichment analysis from eightplex experiment. (XLSX 231 kb)
Rights and permissions
About this article
Cite this article
Phanstiel, D., Brumbaugh, J., Wenger, C. et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Methods 8, 821–827 (2011). https://doi.org/10.1038/nmeth.1699
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1699
This article is cited by
-
Cellular population dynamics shape the route to human pluripotency
Nature Communications (2023)
-
Generation of human A9 dopaminergic pacemakers from induced pluripotent stem cells
Molecular Psychiatry (2022)
-
EGF-activated PI3K/Akt signalling coordinates leucine uptake by regulating LAT3 expression in prostate cancer
Cell Communication and Signaling (2019)
-
DRUGPATH – a novel bioinformatic approach identifies DNA-damage pathway as a regulator of size maintenance in human ESCs and iPSCs
Scientific Reports (2019)
-
An enrichment method based on synergistic and reversible covalent interactions for large-scale analysis of glycoproteins
Nature Communications (2018)