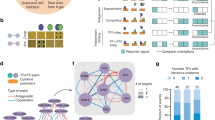
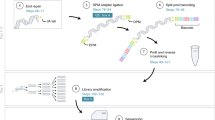
Abstract
Yeast one-hybrid (Y1H) assays provide a gene-centered method for the identification of interactions between gene promoters and regulatory transcription factors (TFs). To date, Y1H assays have involved library screens that are relatively expensive and laborious. We present two Y1H strategies that allow immediate prey identification: matrix assays that use an array of 755 individual Caenorhabditis elegans TFs, and smart-pool assays that use TF multiplexing. Both strategies simplify the Y1H pipeline and reduce the cost of protein-DNA interaction identification. We used a Steiner triple system (STS) to create smart pools of 4–25 TFs. Notably, we uniplexed a small number of highly connected TFs to allow efficient assay deconvolution. Both strategies outperform library screens in terms of coverage, confidence and throughput. These versatile strategies can be adapted both to TFs in other systems and, likely, to other biomolecules and assays as well.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Walhout, A.J.M. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 16, 1445–1454 (2006).
Deplancke, B., Dupuy, D., Vidal, M. & Walhout, A.J.M.A. Gateway-compatible yeast one-hybrid system. Genome Res. 14, 2093–2101 (2004).
Deplancke, B. et al. A gene-centered C. elegans protein-DNA interaction network. Cell 125, 1193–1205 (2006).
Vermeirssen, V. et al. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res.; published online 18 May 2007.
Walhout, A.J.M. et al. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328, 575–592 (2000).
Colbourn, C. & Mathon, R. in The CRC Handbook of Combinatorial Designs. (eds. Colbourn, C. & Dinitz, J.) 66–75 (CRC Press, Boca Raton, Florida, USA, 1996).
Reece-Hoyes, J.S. et al. A compendium of C. elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 6, R110 (2005).
Lamesch, P. et al. C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 14, 2064–2069 (2004).
Wei, C. et al. Closing in on the C. elegans ORFeome by cloning TWINSCAN predictions. Genome Res. 15, 577–582 (2005).
Hartley, J.L., Temple, G.F. & Brasch, M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788–1795 (2000).
Walhout, A.J.M. et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287, 116–122 (2000).
Barrasa, M.I., Vaglio, P., Cavasino, F., Jacotot, L. & Walhout, A.J.M. EDGEdb: a transcription factor-DNA interaction database for the analysis of C. elegans differential gene expression. BMC Genomics 8, 21 (2007).
Jin, F. et al. A pooling-deconvolution strategy for biological network elucidation. Nat. Methods 3, 183–189 (2006).
Thierry-Mieg, N. A new pooling strategy for high-throughput screening: the Shifted Transversal Design. BMC Bioinformatics 7, 28 (2006).
Thierry-Mieg, N. Pooling in systems biology becomes smart. Nat. Methods 3, 161–162 (2006).
Li, S. et al. A map of the interactome network of the metazoan C elegans. Science 303, 540–543 (2004).
Deplancke, B., Vermeirssen, V., Arda, H.E., Martinez, N.J. & Walhout, A.J.M. Gateway-compatible yeast one-hybrid screens. CSH Protocols (doi:10.1101/pdb.prot4590; 2006).
Walhout, A.J.M. & Vidal, M. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24, 297–306 (2001).
Zhong, J., Zhang, H., Stanyon, C.A., Tromp, G. & Finley, R.L., Jr. A strategy for constructing large protein interaction maps using the yeast two-hybrid system: regulated expression arrays and two-phase mating. Genome Res. 13, 2691–2699 (2003).
Reboul, J. et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34, 35–41 (2003).
Hillier, L. & Green, P. OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1, 124–128 (1991).
Acknowledgements
We thank J. Dekker for critical reading of the manuscript; W. Harper (Harvard Medical School) for the Y187 yeast strain; N. Klitgord and M. Vidal (Dana-Farber Cancer Institute) for help with primer design; E. Méndez González (M-3 Informática, S.L.) for helpful suggestions regarding constraint programming and the sequencing staff at Agencourt Bioscience Corporation for technical assistance. This work was supported by BAEF fellowship for Biomedical and Biotechnology Research to V.V., and US National Institute of Diabetes and Digestive and Kidney Diseases grants (DK068429 and DK071713) to A.J.M.W.
Author information
Authors and Affiliations
Contributions
V.V., B.D., J.S.R.-H, H.E.A., C.A.G. and N.J.M. performed all experiments; B.D., M.I.B. and A.J.M.W. conceived the pooling strategy; M.I.B. created the STS design and deconvolution, and performed the bioinformatics analyses. M.R.B. provided TWINSCAN predictions. R.S. and L.D.-S. provided sequencing; V.V., J.S.R.-H. and A.J.M.W. wrote the manuscript.
Ethics declarations
Competing interests
R.S. and L.D.-S. work for Agencourt bioscience.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 2–3, Supplementary Methods. (PDF 876 kb)
Supplementary Table 1
wTF2.1 and clone source and availability. (XLS 515 kb)
Rights and permissions
About this article
Cite this article
Vermeirssen, V., Deplancke, B., Barrasa, M. et al. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat Methods 4, 659–664 (2007). https://doi.org/10.1038/nmeth1063
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1063
This article is cited by
-
MicroRNA-302a is involved in folate deficiency-induced apoptosis through the AKT-FOXO1-BIM pathway in mouse embryonic stem cells
Nutrition & Metabolism (2020)
-
A yeast one‐hybrid and microfluidics‐based pipeline to map mammalian gene regulatory networks
Molecular Systems Biology (2013)
-
What does biologically meaningful mean? A perspective on gene regulatory network validation
Genome Biology (2011)
-
Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping
Nature Methods (2011)