Abstract
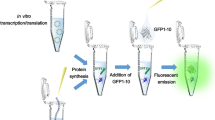
The rapid assessment of protein solubility is essential for evaluating expressed proteins and protein variants for use as reagents for downstream studies. Solubility screens based on antibody blots are complex and have limited screening capacity1,2,3. Protein solubility screens using split β-galactosidase in vivo4 and in vitro5 can perturb protein folding. Split GFP used for monitoring protein interactions folds poorly6, and to overcome this limitation, we recently developed a protein-tagging system based on self-complementing split GFP7 derived from an exceptionally well folded variant of GFP termed 'superfolder GFP'8. Here we present the step-by-step procedure of the solubility assay using split GFP. A 15-amino-acid GFP fragment, GFP 11, is fused to a test protein. The GFP 1–10 detector fragment is expressed separately. These fragments associate spontaneously to form fluorescent GFP. The fragments are soluble, and the GFP 11 tag has minimal effect on protein solubility and folding7. We describe high-throughput protein solubility screens amenable both for in vivo and in vitro formats. The split-GFP system is composed of two vectors used in the same strain: pTET GFP 11 and pET GFP 1–10 (Fig. 1 and Supplementary Note online). The gene encoding the protein of interest is cloned into the pTET GFP 11 vector (resulting in an N-terminal fusion) and transformed into Escherichia coli BL21 (DE3) cells containing the pET GFP 1–10 plasmid. We also describe how this system can be used for selecting soluble proteins from a library of variants (Box 1). The large screening power of the in vivo assay combined with the high accuracy of the in vitro assay point to the efficiency of this two-step split-GFP tool for identifying soluble clones suitable for purification and downstream applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Knaust, R.K. & Nordlund, P. Screening for soluble expression of recombinant proteins in a 96-well format. Anal. Biochem. 297, 79–85 (2001).
Vincentelli, R., Canaan, S., Offant, J., Cambillau, C. & Bignon, C. Automated expression and solubility screening of His-tagged proteins in 96-well format. Anal. Biochem. 346, 77–84 (2005).
Nguyen, H., Martinez, B., Oganesyan, N. & Kim, R. An automated small-scale protein expression and purification screening provides beneficial information for protein production. J. Struct. Funct. Genomics 5, 23–27 (2004).
Wigley, W.C., Stidham, R.D., Smith, N.M., Hunt, J.F. & Thomas, P.J. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat. Biotechnol. 19, 131–136 (2001).
Horecka, J. et al. Antibody-free method for protein detection on blots using enzyme fragment complementation. Biotechniques 40, 381–383 (2006).
Magliery, T.J. et al. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127, 146–157 (2005).
Cabantous, S., Terwilliger, T.C. & Waldo, G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 (2005).
Pedelacq, J.D., Cabantous, S., Tran, T., Terwilliger, T.C. & Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Sambrook, J., Fritsch, E.F. & Maniatis, T., eds. In Molecular Cloning 2nd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1989).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The split GFP and related intellectual properties are the subject of domestic and foreign patent applications by Los Alamos National Laboratories on behalf of the Department of Energy and LANS, L.L.C.
Supplementary information
Rights and permissions
About this article
Cite this article
Cabantous, S., Waldo, G. In vivo and in vitro protein solubility assays using split GFP. Nat Methods 3, 845–854 (2006). https://doi.org/10.1038/nmeth932
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth932
This article is cited by
-
Analysis of protein secretion in Bacillus subtilis by combining a secretion stress biosensor strain with an in vivo split GFP assay
Microbial Cell Factories (2023)
-
Identification and characterization of a novel lytic peptidoglycan transglycosylase (MltC) in Shigella dysenteriae
Brazilian Journal of Microbiology (2023)
-
The iSplit GFP assay detects intracellular recombinant proteins in Bacillus subtilis
Microbial Cell Factories (2021)
-
Scaling production of GFP1-10 detector protein in E. coli for secretion screening by split GFP assay
Microbial Cell Factories (2021)
-
A modular two yeast species secretion system for the production and preparative application of unspecific peroxygenases
Communications Biology (2021)