Abstract
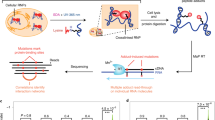
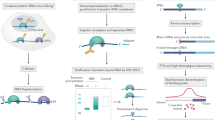
Ribonucleoproteins (RNPs) are key regulators of cellular function. We established an efficient approach, crosslinking of segmentally isotope-labeled RNA and tandem mass spectrometry (CLIR-MS/MS), to localize protein–RNA interactions simultaneously at amino acid and nucleotide resolution. The approach was tested on polypyrimidine tract binding protein 1 and U1 small nuclear RNP. Our method provides distance restraints to support integrative atomic-scale structural modeling and to gain mechanistic insights into RNP-regulated processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Cooper, T.A., Wan, L. & Dreyfuss, G. Cell 136, 777–793 (2009).
Kim, E. et al. Cancer Cell 27, 617–630 (2015).
Leitner, A., Faini, M., Stengel, F. & Aebersold, R. Trends Biochem. Sci. 41, 20–32 (2016).
Kramer, K. et al. Nat. Methods 11, 1064–1070 (2014).
Kwon, S.C. et al. Nat. Struct. Mol. Biol. 20, 1122–1130 (2013).
Beckmann, B.M. et al. Nat. Commun. 6, 10127 (2015).
Lelyveld, V.S., Björkbom, A., Ransey, E.M., Sliz, P. & Szostak, J.W. J. Am. Chem. Soc. 137, 15378–15381 (2015).
Spriggs, K.A., Bushell, M., Mitchell, S.A. & Willis, A.E. Cell Death Differ. 12, 585–591 (2005).
Romanelli, M.G., Diani, E. & Lievens, P.M. Int. J. Mol. Sci. 14, 22906–22932 (2013).
Oberstrass, F.C. et al. Science 309, 2054–2057 (2005).
Han, A. et al. PLoS Comput. Biol. 10, e1003442 (2014).
Kafasla, P. et al. Mol. Cell 34, 556–568 (2009).
Jang, S.K. & Wimmer, E. Genes Dev. 4, 1560–1572 (1990).
Rinner, O. et al. Nat. Methods 5, 315–318 (2008).
Lamichhane, R. et al. Proc. Natl. Acad. Sci. USA 107, 4105–4110 (2010).
Gygi, S.P. et al. Nat. Biotechnol. 17, 994–999 (1999).
Ong, S.E. et al. Mol. Cell. Proteomics 1, 376–386 (2002).
Oda, Y., Huang, K., Cross, F.R., Cowburn, D. & Chait, B.T. Proc. Natl. Acad. Sci. USA 96, 6591–6596 (1999).
Müller, D.R. et al. Anal. Chem. 73, 1927–1934 (2001).
Duss, O., Diarra Dit Konté, N. & Allain, F.H. Methods Enzymol. 565, 537–562 (2015).
Liu, H., Sadygov, R.G. & Yates, J.R. III. Anal. Chem. 76, 4193–4201 (2004).
Daubner, G.M., Cléry, A., Jayne, S., Stevenin, J. & Allain, F.H. EMBO J. 31, 162–174 (2012).
Will, C.L. & Luhrmann, R. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011).
Kondo, Y., Oubridge, C., van Roon, A.M. & Nagai, K. eLife 4, e04986 (2015).
Pomeranz Krummel, D.A., Oubridge, C., Leung, A.K., Li, J. & Nagai, K. Nature 458, 475–480 (2009).
Dorn, G. et al. Mapping protein-RNA interactions with single residue resolution by CLIR-MS/MS. Protocol Exchange http://dx.doi.org/10.1038/protex.2017.031 (2017).
Tugarinov, V., Kanelis, V. & Kay, L.E. Nat. Protoc. 1, 749–754 (2006).
Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2001).
Gasteiger, E. et al. in The Proteomics Protocols Handbook (ed. Walker, J.M.) 571–607 (Humana, Totowa, New Jersey, USA, 2005).
Milligan, J.F., Groebe, D.R., Witherell, G.W. & Uhlenbeck, O.C. Nucleic Acids Res. 15, 8783–8798 (1987).
Kaminski, A. & Jackson, R.J. RNA 4, 626–638 (1998).
Duke, G.M., Hoffman, M.A. & Palmenberg, A.C. J. Virol. 66, 1602–1609 (1992).
Duss, O., Maris, C., von Schroetter, C. & Allain, F.H. Nucleic Acids Res. 38, e188 (2010).
Maniatis, T., Jeffrey, A. & van deSande, H. Biochemistry 14, 3787–3794 (1975).
Sharma, K. et al. Methods 89, 138–148 (2015).
Leitner, A. et al. Anal. Chem. 82, 2726–2733 (2010).
Chambers, M.C. et al. Nat. Biotechnol. 30, 918–920 (2012).
Walzthoeni, T. et al. Nat. Methods 12, 1185–1190 (2015).
Dominguez, C., Schubert, M., Duss, O., Ravindranathan, S. & Allain, F.H. Prog. Nucl. Magn. Reson. Spectrosc. 58, 1–61 (2011).
Lee, W., Revington, M.J., Arrowsmith, C. & Kay, L.E. FEBS Lett. 350, 87–90 (1994).
Peterson, R.D., Theimer, C.A., Wu, H. & Feigon, J. J. Biomol. NMR 28, 59–67 (2004).
Herrmann, T., Güntert, P. & Wüthrich, K. J. Biomol. NMR 24, 171–189 (2002).
Herrmann, T., Güntert, P. & Wüthrich, K. J. Mol. Biol. 319, 209–227 (2002).
Güntert, P. Methods Mol. Biol. 278, 353–378 (2004).
Parisien, M. & Major, F. Nature 452, 51–55 (2008).
Case, D.A. et al. (University of California, San Francisco, 2012).
Vizcaíno, J.A. et al. Nucleic Acids Res. 41, D1063–D1069 (2013).
Acknowledgements
We thank V. Panse for the use of his UV device for crosslinking and N. Diarra dit Konte and O. Duss for advice concerning segmental isotope labeling of RNA. G.D. was supported by a PhD fellowship from the Boehringer Ingelheim Fond. This work was supported by the SNF project 31003A_149921 and the SNF-NCCR RNA and Disease to F.H.-T.A. and by ERC grants Proteomics v3.0 (AdvG grant #233226) and Proteomics4D (AdvG grant #670821) to R.A.
Author information
Authors and Affiliations
Contributions
All authors contributed to the design of experiments, the discussion of the results and the writing of the manuscript. C.v.S. purified isotope-labeled nucleotides. G.D. and A.M. optimized crosslinking conditions. G.D. and S.C. prepared U1snRNP samples. G.D. prepared all other samples and performed and analyzed NMR experiments. A.L. and G.D. performed sample preparation for MS analysis. A.L. performed all LC-MS/MS experiments and analyzed the LC-MS/MS results with support from G.D. J.B. set up the modeling protocol and modeled the RRM2-EMCVF complex based on crosslinking data. G.D. implemented the modeling protocol and modeled the complexes of RRM1, RRM3 and RRM4.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Localization of crosslinks detected for the RRM1-EMCVE complex.
His62 was detected as the sole crosslinking site and is shown in red on the structure of RRM1 in complex with CU-RNA1. The detection of U- and UU-modifications suggests binding of RRM1 to nucleotides 324-326.
Supplementary Figure 2 Segmental labeling of EMCVDElinkF-RNA.
a, strategy for segmental labeling as employed in our study. Notably, only one aliquot of isotope labeled RNA is used independent of the size of the RNA and the number of isotope labeled segments. b, RNAse H digestion of EMCVDElinkF with chimSLD (lane 1), chimLinkF (lane 2), chimSLE (lane 3) and all three chimeras simultaneously (lane 5). The full-length RNA is shown in lane 4, an example for splinted RNA-ligation is shown in lane 6 (input) and 7 (ligation). Note that the isotope labeled RNA fragment is kept as limiting substrate in the reaction.
Supplementary Figure 3 NMR titration experiments for RRM1 (top) and RRM2 (bottom).
(a) EMCVE and (b) EMCVF. The chemical shift perturbations between 1H15N-signals of the free and the bound RRMs indicate binding. The signals of the UCUUU-pentaloop of EMCVF (right) shift upon addition of RRM1 (top) and RRM2 (bottom) due to binding.
Supplementary Figure 4 RRM2 binds preferentially to EMCVF.
a, Upon titration of RRM12 with EMCVF only the RRM2 moiety reaches saturation at a 1:1 ratio (EMCVF:RRM12) and signals overlap almost perfectly with signals of the RRM2-EMCVF complex (yellow). b, In the presence of SLE, RRM1 and RRM2 bind SLE and SLF, respectively, as indicated by the almost perfect superimposition of spectra collected on RRM1-EMCVE (green), RRM2-EMCVF (yellow) and RRM12-EMCVEmutF (blue; right) complexes.
Supplementary Figure 5 Comparison of fingerprint spectra of subcomplexes with their corresponding signals in the PTBP1-EMCVDElinkF complex.
a, the 1H-15N-TROSY signals and, b, the ILV-methyl-1H13C-signals of RRM1 (green), RRM2 (yellow) and RRM34 (magenta) in complex with EMCVE, EMCVF and EMCVDElink, respectively, overlap well with signals of the PTBP1-EMCVDElinkF complex (black) which is typical for identical binding of the RRMs in the subcomplexes.
Supplementary Figure 6 NMR titration of RRM34-EMCVDElink with RRM1.
1H-15N resonances of RRM34-EMCVDElink are not affected by RRM1 addition (color code as indicated). The RRM1-moiety shows the signature of the RRM1-EMCVE complex.
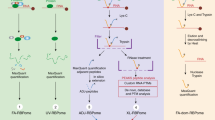
Supplementary Figure 7 Cyana-based modeling strategy using CLIR-MS/MS-derived restraints.
a, peptides and modification sites are identified by CLIR-MS/MS, the mass of the modification infers on its composition as exemplarily shown for RRM3 crosslinks detected in “D”. b, CLIR-MS/MS data are converted to ambiguous upper proton-proton distance limits (illustrated for simplicity as red, dashed lines between carbon atoms) and, c, fed to Cyana 3.02 noeassign. Relevant structural information from published RNA-protein complexes and structure predictions serve to generate complementary ambiguous and unambiguous distance restraints. Calculated atomic resolution models were further refined under amber 12 force-field3. d, representation of the final CLIR-MS/MS restraints on the model of RRM3 in complex with nucleotides 299-306 of SLD.
Supplementary Figure 8 Ensembles of the RRM-RNA complexes modeled using CLIR-MS/MS-derived distance restraints.
Shown are the bundles of the 20 final structures of the, a, RRM1-EMCVE, b, RRM2-EMCVF, c, RRM3-EMCVD and, d, RRM4-EMCVlink complex. Only nucleotides 354-364, 300-305 and 340-344 are displayed in A, C and D, respectively, as these represent the well-defined binding site. RMSD values (Deviation of coordinates to their mean) are calculated for amino acids 47-153 (RRM1), 181-244 and 255-273 (RRM2) and 336-531 (RRM34 tandem domain) and nucleotides 321-336 (EMCVE), 354-364 (EMCVF), 302-305 (EMCVD) and 341-344 (EMCVlink).
Supplementary Figure 9 Recognition of the EMCV-IRES domains D–F by PTBP1-RRMs according to CLIR-MS/MS-based models.
Hydrophobic and specific hydrogen bond interactions are depicted for each binding pocket of the RRMs as derived from the CLIR-MS/MS-based models and compared with the published interactions detected in an RRM1-stemloop complex (pdb: 3N2O), the NMR derived model of RRM2-EMCVF and in the complex of RRM34 with ssRNA1. Recognition of nucleotides in the reference structures is shown in gray if different. Please note that U360 is looped out and not recognized by RRM2.
Supplementary Figure 10 Comparison of the RRM2-EMCVF models derived from NMR and CLIR-MS/MS.
RRM2 recognizes EMCVF by contacting C358 and U359 in pocket 1 and 2 by base-specific hydrogen bonds in both, a, NMR and, b, CLIR-MS/MS derived models. While U360 does not contact RRM2, U361 stacks on Tyr 267.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 1–3 and Supplementary Notes 1–6 (PDF 2178 kb)
Supplementary Table 4
Modeling restraints (XLSX 519 kb)
Supplementary Data 1
CLIR-MS/MS derived model of RRM1 bound to stem loop E (ZIP 798 kb)
Supplementary Data 2
CLIR-MS/MS derived model of RRM2 bound to stem loop F (ZIP 931 kb)
Supplementary Data 3
CLIR-MS/MS derived model of RRM3 bound to stem loop D (ZIP 1175 kb)
Supplementary Data 4
CLIR-MS/MS derived model of RRM4 bound to linker sequence E-F (ZIP 1206 kb)
Supplementary Data 5
NMR derived model of RRM2 bound to stem loop F (ZIP 1008 kb)
Supplementary Protocol
Mapping protein-RNA interactions with single residue resolution by CLIR-MS/MS (PDF 389 kb)
Source data
Rights and permissions
About this article
Cite this article
Dorn, G., Leitner, A., Boudet, J. et al. Structural modeling of protein–RNA complexes using crosslinking of segmentally isotope-labeled RNA and MS/MS. Nat Methods 14, 487–490 (2017). https://doi.org/10.1038/nmeth.4235
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4235
This article is cited by
-
Integrative solution structure of PTBP1-IRES complex reveals strong compaction and ordering with residual conformational flexibility
Nature Communications (2023)
-
Probing the secrets of probes
Nature Chemical Biology (2022)
-
Nucleotide-amino acid π-stacking interactions initiate photo cross-linking in RNA-protein complexes
Nature Communications (2022)
-
Photoactivatable ribonucleosides mark base-specific RNA-binding sites
Nature Communications (2021)
-
NMR and EPR reveal a compaction of the RNA-binding protein FUS upon droplet formation
Nature Chemical Biology (2021)