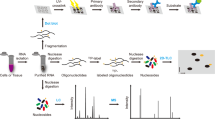
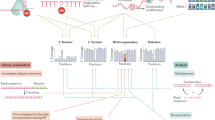
Abstract
In recent years, major breakthroughs in RNA-modification-mediated regulation of gene expression have been made, leading to the emerging field of epitranscriptomics.Our understanding of the distribution, regulation and function of these dynamic RNA modifications is based on sequencing technologies. In this Review, we focus on the major mRNA modifications in the transcriptome of eukaryotic cells: N6-methyladenosine, N6, 2′-O-dimethyladenosine, 5-methylcytidine, 5-hydroxylmethylcytidine, inosine, pseudouridine and N1-methyladenosine. We discuss the sequencing technologies used to profile these epitranscriptomic marks, including scale, resolution, quantitative feature, pre-enrichment capability and the corresponding bioinformatics tools. We also discuss the challenges of epitranscriptome profiling and highlight the prospect of future detection tools. We aim to guide the choice of different detection methods and inspire new ideas in RNA biology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
10 February 2017
In the version of this article initially published, author affiliation numbers were incorrect. Xiaoyu Li originally had affiliation 1; this has been changed to affiliations 1 and 2. Xushen Xiong originally had affiliations 1 and 2; these have been changed to affiliations 1–3. Chengqi Yi originally had affiliations 1 and 3; these have been changed to affiliations 2 and 4. The error has been corrected in the HTML and PDF versions of the article.
References
Machnicka, M.A. et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
He, C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 6, 863–865 (2010). This paper termed the scope and mechanisms of dynamic RNA modifications 'RNA epigenetics' for the first time.
Saletore, Y. et al. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175 (2012). This paper first termed the multitude of RNA modifications as 'epitranscriptome'.
Fu, Y., Dominissini, D., Rechavi, G. & He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014).
Liu, N. & Pan, T. N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 23, 98–102 (2016).
Gilbert, W.V., Bell, T.A. & Schaening, C. Messenger RNA modifications: form, distribution, and function. Science 352, 1408–1412 (2016).
Meyer, K.D. & Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014).
Lee, M., Kim, B. & Kim, V.N. Emerging roles of RNA modification: m(6)A and U-tail. Cell 158, 980–987 (2014).
Saletore, Y., Chen-Kiang, S. & Mason, C.E. Novel RNA regulatory mechanisms revealed in the epitranscriptome. RNA Biol. 10, 342–346 (2013).
Bokar, J.A., Rath-Shambaugh, M.E., Ludwiczak, R., Narayan, P. & Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269, 17697–17704 (1994).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Bokar, J.A., Shambaugh, M.E., Polayes, D., Matera, A.G. & Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997).
Ping, X.L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Reports 8, 284–296 (2014).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). This work discovered the first m6A RNA demethylase ('eraser')—FTO, demonstrating reversible RNA methylation in the human transcriptome.
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Alarcón, C.R. et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). This is an important study that reports the first m6A methylome in the mammalian transcriptome.
Liu, N. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 (2015).
Meyer, K.D. et al. 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Wang, X. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Xiao, W. et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Lichinchi, G. et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011 (2016).
Tirumuru, N. et al. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 5, e15528 (2016).
Gokhale, N.S. et al. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016).
Kennedy, E.M. et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19, 675–685 (2016).
Lichinchi, G. et al. Dynamics of human and viral RNA methylation during zika virus infection. Cell Host Microbe 20, 666–673 (2016).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 71, 3971–3975 (1974).
Csepany, T., Lin, A., Baldick, C.J. Jr. & Beemon, K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 265, 20117–20122 (1990).
Narayan, P. & Rottman, F.M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science 242, 1159–1162 (1988).
Narayan, P., Ludwiczak, R.L., Goodwin, E.C. & Rottman, F.M. Context effects on N6-adenosine methylation sites in prolactin mRNA. Nucleic Acids Res. 22, 419–426 (1994).
Meyer, K.D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012). The other study that first reports the transcriptome-wide m6A methylome.
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013).
Chen, K. et al. High-resolution N(6) -methyladenosine (m(6)A) map using photo-crosslinking-assisted m(6)A sequencing. Angew. Chem. Int. Edn Engl. 54, 1587–1590 (2015).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Molinie, B. et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13, 692–698 (2016).
Schibler, U. & Perry, R.P. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Res. 4, 4133–4149 (1977).
Keith, J.M., Ensinger, M.J. & Mose, B. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 253, 5033–5039 (1978).
Wei, C., Gershowitz, A. & Moss, B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253 (1975).
Munns, T.W., Oberst, R.J., Sims, H.F. & Liszewski, M.K. Antibody-nucleic acid complexes. Immunospecific recognition of 7-methylguanine- and N6-methyladenine-containing 5′-terminal oligonucleotides of mRNA. J. Biol. Chem. 254, 4327–4330 (1979).
Motorin, Y. & Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944 (2010).
Squires, J.E. & Preiss, T. Function and detection of 5-methylcytosine in eukaryotic RNA. Epigenomics 2, 709–715 (2010).
Chow, C.S., Lamichhane, T.N. & Mahto, S.K. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2, 610–619 (2007).
Brzezicha, B. et al. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 34, 6034–6043 (2006).
Goll, M.G. et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 (2006).
Schaefer, M., Pollex, T., Hanna, K. & Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12 (2009).
Squires, J.E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Hussain, S., Aleksic, J., Blanco, S., Dietmann, S. & Frye, M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 14, 215 (2013).
Shafik, A., Schumann, U., Evers, M., Sibbritt, T. & Preiss, T. The emerging epitranscriptomics of long noncoding RNAs. Biochim. Biophys. Acta 1859, 59–70 (2016).
Edelheit, S., Schwartz, S., Mumbach, M.R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602 (2013).
Khoddami, V. & Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31, 458–464 (2013).
Hussain, S. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Reports 4, 255–261 (2013).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Li, S. & Mason, C.E. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 15, 127–150 (2014).
Rácz, I., Király, I. & Lásztily, D. Effect of light on the nucleotide composition of rRNA of wheat seedlings. Planta 142, 263–267 (1978).
Delatte, B. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Fu, L. et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 136, 11582–11585 (2014).
Huber, S.M. et al. Formation and abundance of 5-hydroxymethylcytosine in RNA. ChemBioChem 16, 752–755 (2015).
Booth, M.J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 (2002).
Jepson, J.E. & Reenan, R.A. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta 1779, 459–470 (2008).
Levanon, E.Y. et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005 (2004).
Athanasiadis, A., Rich, A. & Maas, S. Widespread A-to-I RNA editing of alu-containing mRNAs in the human transcriptome. PLoS Biol. 2, e391 (2004).
Wulff, B.E., Sakurai, M. & Nishikura, K. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat. Rev. Genet. 12, 81–85 (2011).
Kim, D.D. et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 14, 1719–1725 (2004).
Li, J.B. et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324, 1210–1213 (2009).
Li, M. et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333, 53–58 (2011).
Lin, W., Piskol, R., Tan, M.H. & Li, J.B. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335, 1302-e (2012).
Pickrell, J.K., Gilad, Y. & Pritchard, J.K. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335, 1302-d (2012).
Kleinman, C.L. & Majewski, J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335, 1302-c (2012).
Ju, Y.S. et al. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat. Genet. 43, 745–752 (2011).
Bahn, J.H. et al. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 22, 142–150 (2012).
Ramaswami, G. et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods 9, 579–581 (2012).
Peng, Z. et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30, 253–260 (2012).
Ramaswami, G. et al. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 10, 128–132 (2013).
Sakurai, M., Yano, T., Kawabata, H., Ueda, H. & Suzuki, T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 6, 733–740 (2010).
Sakurai, M. et al. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 24, 522–534 (2014).
Karijolich, J., Yi, C. & Yu, Y.T. Transcriptome-wide dynamics of RNA pseudouridylation. Nat. Rev. Mol. Cell Biol. 16, 581–585 (2015).
Kiss, T., Fayet-Lebaron, E. & Jády, B.E. Box H/ACA small ribonucleoproteins. Mol. Cell 37, 597–606 (2010).
Hamma, T. & Ferré-D'Amaré, A.R. Pseudouridine synthases. Chem. Biol. 13, 1125–1135 (2006).
Li, X., Ma, S. & Yi, C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 33, 108–116 (2016).
Decatur, W.A. & Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002).
Baudin-Baillieu, A. et al. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 37, 7665–7677 (2009).
Jack, K. et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44, 660–666 (2011).
Gilham, P.T. An addition reaction specific for uridine and guanosine nucleotides and its application to the modification of ribonuclease action. J. Am. Chem. Soc. 84, 687–688 (1962).
Bakin, A. & Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762 (1993).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014). This work reports Ψ-seq to map Ψ sites in the yeast and human transcriptome.
Carlile, T.M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014). This study reports Pseudo-seq to map Ψ sites in the yeast and human transcriptome.
Lovejoy, A.F., Riordan, D.P. & Brown, P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015). This study reports CeU-Seq for the comprehensive identification of Ψ sites in the human and mouse transcriptome.
Zaringhalam, M. & Papavasiliou, F.N. Pseudouridylation meets next-generation sequencing. Methods 107, 63–72 (2016).
Kellner, S., Burhenne, J. & Helm, M. Detection of RNA modifications. RNA Biol. 7, 237–247 (2010).
Behm-Ansmant, I., Helm, M. & Motorin, Y. Use of specific chemical reagents for detection of modified nucleotides in RNA. J. Nucleic Acids 2011, 408053 (2011).
Ozanick, S., Krecic, A., Andersland, J. & Anderson, J.T. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11, 1281–1290 (2005).
Chujo, T. & Suzuki, T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 18, 2269–2276 (2012).
Schevitz, R.W. et al. Crystal structure of a eukaryotic initiator tRNA. Nature 278, 188–190 (1979).
Vilardo, E. et al. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 40, 11583–11593 (2012).
Hauenschild, R. et al. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 43, 9950–9964 (2015).
Waku, T. et al. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J. Cell Sci. 129, 2382–2393 (2016).
Dominissini, D. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016). This study reports the transcriptome-wide m1A methylome for the first time.
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016). This work reports the transcriptome-wide m1A methylome for the first time.
Garalde, D.R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Preprint at http://biorxiv.org/content/early/2016/08/12/068809 (2016).
Dong, Z.W. et al. RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 40, e157 (2012).
Lacoux, C. et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 40, 4086–4096 (2012).
Birkedal, U. et al. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Edn Engl. 54, 451–455 (2015).
Gumienny, R., Jedlinski, D., Martin, G., Vina-Villaseca, A. & Zavolan, M. High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq. Preprint at http://biorxiv.org/content/early/2016/01/19/037259 (2016).
Incarnato, D. et al. High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. Nucleic Acids Res. Sep 9, gkw810 (2016).
Marchand, V., Blanloeil-Oillo, F., Helm, M. & Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 44, e135 (2016).
Acknowledgements
We thank all members of the Yi laboratory for their insights and discussions. This work was supported by the National Basic Research Foundation of China (grant nos. MOST2016YFC0900300 and 2014CB964900) and the National Natural Science Foundation of China (grant no. 21522201).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Table
Supplementary Table 1 (PDF 109 kb)
Rights and permissions
About this article
Cite this article
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods 14, 23–31 (2017). https://doi.org/10.1038/nmeth.4110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4110
This article is cited by
-
Research progress of N1-methyladenosine RNA modification in cancer
Cell Communication and Signaling (2024)
-
Clinical significance of RNA methylation in hepatocellular carcinoma
Cell Communication and Signaling (2024)
-
Age-related noncanonical TRMT6–TRMT61A signaling impairs hematopoietic stem cells
Nature Aging (2024)
-
m6A/m1A/m5C-Associated Methylation Alterations and Immune Profile in MDD
Molecular Neurobiology (2024)
-
Regulation and functions of non-m6A mRNA modifications
Nature Reviews Molecular Cell Biology (2023)