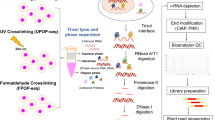
Abstract
As RNA-binding proteins (RBPs) play essential roles in cellular physiology by interacting with target RNA molecules, binding site identification by UV crosslinking and immunoprecipitation (CLIP) of ribonucleoprotein complexes is critical to understanding RBP function. However, current CLIP protocols are technically demanding and yield low-complexity libraries with high experimental failure rates. We have developed an enhanced CLIP (eCLIP) protocol that decreases requisite amplification by ∼1,000-fold, decreasing discarded PCR duplicate reads by ∼60% while maintaining single-nucleotide binding resolution. By simplifying the generation of paired IgG and size-matched input controls, eCLIP improves specificity in the discovery of authentic binding sites. We generated 102 eCLIP experiments for 73 diverse RBPs in HepG2 and K562 cells (available at https://www.encodeproject.org), demonstrating that eCLIP enables large-scale and robust profiling, with amplification and sample requirements similar to those of ChIP-seq. eCLIP enables integrative analysis of diverse RBPs to reveal factor-specific profiles, common artifacts for CLIP and RNA-centric perspectives on RBP activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Gerstberger, S., Hafner, M. & Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 (2014).
Castello, A., Fischer, B., Hentze, M.W. & Preiss, T. RNA-binding proteins in Mendelian disease. Trends. Genet. 29, 318–327 (2013).
Nussbacher, J.K., Batra, R., Lagier-Tourenne, C. & Yeo, G.W. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 38, 226–236 (2015).
Ule, J. et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
König, J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 (2010).
Shishkin, A.A. et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015).
Huppertz, I. et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65, 274–287 (2014).
Yeo, G.W. et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16, 130–137 (2009).
Darnell, J.C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Lovci, M.T. et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 20, 1434–1442 (2013).
Weyn-Vanhentenryck, S.M. et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. http://dx.doi.org/10.1016/j.celrep.2014.02.005 (2014).
Brooks, L. III et al. A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. RNA 21, 1943–1965 (2015).
Reyes-Herrera, P.H., Speck-Hernandez, C.A., Sierra, C.A. & Herrera, S. BackCLIP: a tool to identify common background presence in PAR-CLIP datasets. Bioinformatics (2015).
Friedersdorf, M.B. & Keene, J.D. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 15, R2 (2014).
Tenenbaum, S.A., Carson, C.C., Lager, P.J. & Keene, J.D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 97, 14085–14090 (2000).
Rozowsky, J. et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 27, 66–75 (2009).
Li, Q., Brown, J.B., Huang, H. & Bickel, P.J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011).
Sundararaman, B. et al. Resources for the comprehensive discovery of functional RNA elements. Mol. Cell http://dx.doi.org/10.1016/j.molcel.2016.02.012 (2016).
Richman, T.R. et al. A bifunctional protein regulates mitochondrial protein synthesis. Nucleic Acids Res. 42, 5483–5494 (2014).
Grainger, R.J. & Beggs, J.D. Prp8 protein: at the heart of the spliceosome. RNA 11, 533–557 (2005).
Rappsilber, J., Ajuh, P., Lamond, A.I. & Mann, M. SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J. Biol. Chem. 276, 31142–31150 (2001).
Rackham, O., Mercer, T.R. & Filipovska, A. The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. Wiley Interdiscip. Rev. RNA. 3, 675–695 (2012).
Matera, A.G. & Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014).
Krueger, B.J. et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36, 2219–2229 (2008).
McHugh, C.A. et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015).
Chu, C. et al. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015).
Royce-Tolland, M.E. et al. The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nat. Struct. Mol. Biol. 17, 948–954 (2010).
Guo, F. et al. Regulation of MALAT1 expression by TDP43 controls the migration and invasion of non-small cell lung cancer cells in vitro. Biochem. Biophys. Res. Commun. 465, 293–298 (2015).
Tripathi, V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010).
Acknowledgements
The authors would like to thank members of the Yeo lab (particularly S. Aigner and S. Markmiller) as well as colleagues J. Van Nostrand, Y. Kobayashi, B.R. Graveley and C.B. Burge for critical reading of the manuscript, and M. Blanco with early method development. This work was supported by grants from the US National Institutes of Health (HG004659, U54HG007005 and NS075449 to G.W.Y.), and by the US National Institutes of Health Director's Early Independence Award (DP5OD012190) and funds from the California Institute of Technology to M.G. We would also like to thank Ionis Pharmaceuticals for sharing reagents. E.L.V.N. is a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2172-13). G.W.Y. is an Alfred P. Sloan Research Fellow. G.A.P. is supported by the National Science Foundation Graduate Research Fellowship.
Author information
Authors and Affiliations
Contributions
E.L.V.N., A.A.S., M.G., and G.W.Y. conceived the study. E.L.V.N., A.A.S., and C.S. developed the eCLIP methodology. E.L.V.N., C.G.-B., and S.M.B. performed 293T eCLIP and RBFOX2 knockdown experiments. F.R. provided antisense oligonucleotides (ASOs) and M.Y.F. performed ASO experiments. C.G.-B., B.S., S.M.B., T.B.N., K.E., and R.S. performed K562 and HepG2 eCLIP experiments. E.L.V.N. and G.A.P. performed computational analyses. E.L.V.N. and G.W.Y. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
F.R. is a paid employee of Ionis Pharmaceuticals.
Integrated supplementary information
Supplementary Figure 1 Large-scale iCLIP experiments indicate poor efficiency.
(a) The fraction of usable (non-PCR duplicate, uniquely mapping) reads out of uniquely mapped reads is shown for 279 published CLIP experiments: 127 iCLIP (12 performed for the ENCODE consortium as well as 115 published) and 152 other (including PAR-CLIP and HITS-CLIP). Datasets and read-level processing statistics are listed in Supplementary Table 1. Histogram indicates the number of CLIP experiments within the indicated usable fraction bin. (b) Out of 66 iCLIP experiments performed for the ENCODE consortium, only 15 showed successful amplification of library in both biological replicates (all requiring 24-32 cycles of PCR).
Supplementary Figure 2 Optional sample pooling strategy and eCLIP computational analysis workflow.
(a) At the 3′ RNA adapter ligation step in eCLIP, the RNA adapter includes a barcode sequence, enabling pooling of multiple experiments before the protein gel electrophoresis step. Note that pooled samples must have identical desired cut size on the nitrocellulose membrane, and should have a similar number of RNA molecules (to avoid over- or under-sequencing of individual experiments within the pooled sample). (b) Schematic of eCLIP computational analysis pipeline. Squares indicate processing steps, with processing output used for downstream analyses indicated as filled green circles. Software packages used are indicated in bold.
Supplementary Figure 3 eCLIP of RBFOX2 improves library efficiency over iCLIP.
(a) eCLIP and iCLIP were performed using the same RBFOX2 antibody on HEK293T cells. (b) Western blot of RBFOX2 immunoprecipitation during eCLIP. Replicates (Rep1 and Rep2) were performed on ‘biological replicate’ 293T samples grown and crosslinked ~2 months apart. Red dotted line indicates region excised for eCLIP library preparation. (c) Western blot of SLBP immunoprecipitation during eCLIP, performed with two concentrations (5U or 40U) of RNase I during fragmentation. (d) eCLIP requires decreased amplification compared to iCLIP. To more easily compare across samples, we defined an extrapolated cycle number (eCT) as the number of cycles needed to obtain 100 fmoles of amplified material, extrapolated from the final library volume, final library concentration, and number of PCR cycles done, assuming doubling at each cycle. (e) Fraction of reads that uniquely map to the genome is similar between iCLIP and eCLIP. (f) Peak locations (top) and de novo motifs identified by HOMER (middle) show similar signal between iCLIP and eCLIP. Proximal intron indicates the region ≤ 500 nt from the 5′ or 3′ splice site, with the remainder annotated as distal intron. Motifs were identified relative to a background of randomly selected regions from the same annotation class (e.g. CDS exons, proximal introns, etc). Significance indicated is as reported by HOMER. The subset of clusters significantly enriched vs SMInput (≥ 8-fold, p ≤ 10-5 by Fisher Exact or Chi Square test) show increased intronic localization for both eCLIP replicates (bottom).
Supplementary Figure 4 eCLIP improves library efficiency over iCLIP for IGF2BP1 and IGF2BP2.
(a-b) eCT shows > 10 cycle improvement for eCLIP over iCLIP of (a) IGF2BP1 and (b) IGF2BP2 in K562 cells. (c-d) eCLIP shows improvement in the fraction of uniquely mapped reads that are usable relative to iCLIP when identical numbers of reads are downsampled from biological replicates of (c) IGF2BP1 and (d) IGF2BP2 in K562 cells. Correlation to regression (R2) is indicated, where * indicates best fit by logarithmic regression and unlabeled indicates linear regression.
Supplementary Figure 5 Reverse transcriptase termination at crosslink sites leaves stereotypical motif frequencies flanking eCLIP sequence reads.
(a-d) Plots indicate the enrichment of indicated motifs at each position flanking the start position of mapped reads for (a) RBFOX2 eCLIP and iCLIP in 293T cells, (b) TARDBP eCLIP in K562 cells, (c) PUM2 eCLIP in K562 cells, and (d) TRA2A eCLIP in HepG2 and K562 cells. For each dataset, the frequency of the indicated kmer at each position was tallied, and compared against the frequency in paired SMInput to obtain single-nucleotide enrichment profiles. (a) RBFOX2 shows enrichment for crosslinking at G2 and G6 positions in both iCLIP and eCLIP, consistent with previous results. (b) TARDBP single-nucleotide profile indicates enrichment for the GAAUG at –8 and –4 nucleotides relative to read start positions. (c) PUM2 indicates UGUANAUA motif at –2, –1, and 0 relative to read start positions. (d) For TRA2A, the canonical GAAGAA motif is highly enriched around read starts but does not show specific fold-enrichment at any particular position, indicating that the majority of termination-inducing crosslinks occur at positions within the RNA that are distant from the sequence-specific site of TRA2A interaction.
Supplementary Figure 6 Functional validation of eCLIP binding sites by antisense oligonucleotide (ASO) blocking.
(a) Tracks indicate read density for iCLIP and eCLIP of RBFOX2 at an RBFOX2 binding site flanking exon 9 of NDEL1, and the location of three antisense oligonucleotides (with uniform 2′-O-methoxyethyl-modified nucleotides and a phosphorothioate backbone). Darkened bars underneath indicate peaks significantly enriched above SMInput. Read density tracks are normalized to show the number of reads per million total usable reads (RPM). (b) Treatment of 293T cells with NDEL1-targeting and control ASOs indicates that blocking RBFOX2 binding increases cassette exon exclusion. Asterisks denote significance determined by Student’s t-Test performed on the change in percent spliced in (ΔΨ). (c-f) Similar analysis indicates ASO blocking of RBFOX2 binding affects splicing of (c-d) ECT2 exon 5 and (e-f) EPB41 exon 16.
Supplementary Figure 7 Validation of standard eCLIP conditions across fragmentation conditions.
(a) Histone RNA read density is increased in 5 U RNase I digestion relative to 40 U, but both are dramatically increased above SMInput, RBFOX2, and IgG controls. See Supplementary Data for histone gene list. (b) eCLIP of SLBP shows similar enrichment at HIST1H1C 3′UTR with 40 U or 5 U of RNase I fragmentation. (c-f) Multiple analyses indicate similar eCLIP results across a range of 0-2000U RNase I fragmentation conditions for RBFOX2 eCLIP in 293T cells (c) Increased fragmentation (by increased RNase I concentration) slightly increases the fraction of intronic RBFOX2 signal relative to exonic, but intronic regions compromise the majority of bases covered across all conditions. Stacked bars indicate the fraction of bases covered by RBFOX2 clusters (identified by CLIPper) with respect to the indicated RNA transcript regions. (d) Bar graphs indicate the number of clusters identified in RBFOX2 eCLIP fragmentation experiments. Most showed 20,000-40,000 clusters, with the exception of the 2000U condition in which only 1,137 clusters were identified. (e) Read density tracks show eCLIP binding profiles flanking an RBFOX2-dependent cassette exon in EPB41L2. With the exception of the 2000 U condition, conditions show similar enrichment patterns and RPM coverage. (f) RBFOX2 motif (UGCAUG) enrichment in CLIPper-identified clusters increases with increasing RNase I fragmentation. Fold-enrichment shown is relative to frequency observed in ten random permutations of cluster sequences.
Supplementary Figure 8 Paired Size-Matched Input (SMInput) reveals enrichment over common background in CLIP of histone-binding SLBP.
(a) At an abundantly expressed housekeeping gene EEF2 (Eukaryotic Translation Elongation Factor 2), similar read density is observed in eCLIP of histone-binding SLBP as in SMInput, indicating that this signal is not indicative of true binding events. Tracks below read density indicate CLIPper clusters, with darkened clusters indicating clusters significantly enriched above SMInput. Below, exonic-binding LIN28B shows significant binding to exon 5 of EEF2, indicating that enriched binding events can be observed above this background. (b) SLBP eCLIP shows specific enrichment for reads in histone coding exon (CDS, circles) and 3′UTR (square) regions relative to paired SMInput. Each point indicates a gene, with the x-position indicating the number of reads observed in SMInput (plus a pseudocount of 1) and the y-position indicating the fold-enrichment in SLBP 293T eCLIP (Rep1). Histone genes are indicated in pink. Significantly enriched regions (fold-enrichment ≥ 4-fold, p-value ≤ 10-5 in eCLIP vs SMInput) are indicated by open shapes (Significance is determined by Yates’ Chi-Square test, with Fisher’s Exact tests when eCLIP or SMInput has < 5 reads). (c-d) Read density (normalized as reads per million (RPM)) is shown for eCLIP of histone processing factor SLBP, along with paired SMInput, for SLBP-enriched target HIST1H1C and non-enriched U12 snRNA transcript RNU12. Rectangles below SLBP read density track indicate clusters identified with the CLIPper peak identification algorithm, with fold-enrichment in eCLIP indicated below. (e) All CLIPper-identified clusters identified for SLBP 293T eCLIP (Rep1) are plotted based on their fold-enrichment and significance compared to paired SMInput. Significance is determined by Yates’ Chi-Square test, with Fisher’s Exact tests (minimum p-value = 2.2 × 10-16) when eCLIP or SMInput has < 5 reads. Only 284 clusters (1.2%) are enriched at least 8-fold with p ≤ 10-5 by Fisher Exact or Chi Square test in eCLIP (pink shaded box). Clusters overlapping histone genes (indicated in pink) are shifted towards high significance and fold-enrichment.
Supplementary Figure 9 Paired Size-Matched Input (SMInput) reveals enrichment over common background in CLIP of splicing regulator RBFOX2.
(a) All CLIPper-identified clusters identified for RBFOX2 293T eCLIP (Rep1) are plotted based on their fold-enrichment and significance compared to paired SMInput. Only 5,954 clusters (7.9%) are enriched at least 8-fold with p ≤ 10-5 by Fisher Exact or Chi Square test in eCLIP (green shaded box). Clusters overlapping introns flanking a set of 197 exons with RBFOX2 dependent splicing observed from microarray analysis of RBFOX2 knockdown (shRNA 1; Supplementary Fig. 10a-f) are indicated in green. (b) The subset of 50,853 RBFOX2 eCLIP clusters with either pre-normalized (CLIPper) or SMInput normalized p-value ≤ 10-5 were ranked by pre-normalized CLIPper p-value (left) or by SMInput normalization (right), as in Figure 2D. (Center) for clusters located in introns flanking RBFOX2-dependent cassette exons (Supplementary Fig. 10), change in rank is indicated by green lines, with significance determined by Kolmogorov-Smirnov test. Histograms indicate the number of RBFOX2-dependent cassette exon-flanking binding sites in each bin for clusters sorted by (left) CLIPper p-value, or (right) SMInput-normalized p-value. (c) Points indicate the enrichment for the RBFOX2 (UGCAUG) motif in each bin for RBFOX2 eCLIP clusters ranked by SMInput fold-enrichment (green) or pre-normalized CLIPper p-value (grey), with Replicate 1 indicated as solid and Replicate 2 as dashed lines. SMInput normalization decreases the frequency of motifs at non- or lowly-enriched clusters (left; indicating down-ranking of false positive clusters), but increases the frequency of motifs at highly enriched clusters (right; indicating up-ranking of true positive clusters). Motif enrichment was determined by counting the number of UGCAUG 6-mers in cluster sequences, and in 10 random permutations of the sequence within each clusters. (d) For the data shown in C, clusters were separated into two bins: ‘depleted’ clusters with decreased RPM in eCLIP vs SMInput, and ‘significantly enriched’ clusters with eCLIP read density at least 8-fold enriched and p ≤ 10-5 relative to SMInput. For both all CLIPper clusters (black), as well as a more stringent subset of only those with CLIPper p-value ≤ 10-5 (grey), depleted clusters show little or no enrichment for RBFOX2 motifs, whereas significantly enriched peaks show > 20-fold enrichment.
Supplementary Figure 10 Splicing-sensitive microarray analysis identifies RBFOX2-dependent cassette exons.
(a) RBFOX2 knockdown by transduction and selection for shRNA was performed in 293T cells, with splicing profiled by Affymetrix HTA2.0 microarray. Each knockdown was performed in biological triplicate, and each sample was separately prepared and hybridized. (B-C) Validation of RBFOX2 knockdown by western blot for shRNA 1 (TRCN0000074544), shRNA 2 (TRCN0000074546), and shRNA 3 (TRCN0000074543). (b) After lentiviral infection and puromycin selection, 293T cells were lysed in eCLIP lysis buffer, run on standard NuPAGE Novex 4-12% Bis-Tris gel (Thermo Fisher), transferred to PVDF membrane, and imaged on a LiCor Odyssey using RBFOX2 (rabbit A300-864A, Bethyl) and GAPDH (mouse ab8245, Abcam) primary and fluorescent secondary antibodies. (c) Band intensity was quantitated using LiCor ImageStudio Lite software. (d-f) Analysis of splicing-sensitive microarrays identifies RBFOX2-dependent cassette exons. (d) (top) Probes corresponding to cassette exon inclusion (AS exon probes (purple) and UP and DN junction probes (red)) and exclusion (AS junction (green)) were identified for all cassette exons profiled on the array. (bottom left) All probes for each gene were then normalized across samples to obtain residuals. (bottom right) Change in splicing is quantified by calculating a SepScore, defined as the mean residual signal for exclusion probes minus the mean signal for inclusion probes. (e) Heatmap indicates SepScore across all nine knockdown samples (relative to the average of non-target control samples) for the set of 299 events that showed significant change in either inclusion or exclusion probes (p ≤ 0.001 by t-Test), as well as |SepScore| ≥ 0.5 for at least one shRNA. Comparison of events significant in any of the three knockdown experiments showed high similarity in splicing change across shRNAs. (f) Splicing analysis SepScore shows increased exclusion for NDEL1 exon 9, ECT2 exon 5, and EPB41 exon 16 upon RBFOX2 knockdown by shRNA.
Supplementary Figure 11 SMInput-normalization distinguishes significantly enriched eCLIP peaks which contain known binding motifs from clusters depleted in eCLIP which lack motifs.
(a-c) As shown in Supplementary Figure 9D for RBFOX2, clusters for eCLIP of (a) TARDBP, (b) PUM2, and (c) TRA2A were separated into two bins: ‘depleted’ clusters with decreased RPM in eCLIP vs SMInput, and ‘significantly enriched’ clusters with eCLIP read density at least 8-fold enriched and p ≤ 10-5 relative to SMInput. Shown are motif enrichment for all CLIPper clusters (black), as well as a more stringent subset of only those with CLIPper p-value ≤ 10-5 (grey).
Supplementary Figure 12 eCLIP shows high reproducibility across biological replicates.
(a) SLBP eCLIP fold-enrichment over SMInput at histone RNAs is reproducible across biological replicate experiments. Each point indicates eCLIP fold-enrichment over paired SMInput for the CDS (circle) or 3′UTR (square) of genes profiled in independent biological replicate SLBP eCLIP experiments. Histone genes are indicated in pink, with open circles indicating significantly enriched regions (fold-enrichment ≥ 4-fold, p-value ≤ 10-5 in eCLIP vs SMInput). Both CDS (R2 = 0.50) and 3′UTR (R2 = 0.73) show significant correlation (p < 10-300, all significance determined by standard conversion of r values to t-statistic), and show enrichment at most histones. (b) SLBP clusters were identified in Replicate 1, and for each cluster the fold-enrichment was determined for both Replicate 1 and Replicate 2 eCLIP. Histone-overlapping points are indicated in pink, with significantly enriched peaks indicated in blue. Attached histograms show the number of significantly-enriched peaks with specified fold-enrichment in Replicate 1 (top) and Replicate 2 (right). (c) Correlation in read density across biological replicate RBFOX2 eCLIP experiments. Clusters were first identified in Replicate 1, and then each point indicates RBFOX2 eCLIP RPM for Rep1 and Rep2 at these clusters. (d) SMInput-normalized eCLIP peak signal shows high correlation between biological replicate RBFOX2 (and SMInput) experiments. Clusters are identified using CLIPper on Rep1 only, and points indicate fold-enrichment in eCLIP over SMInput for these regions across biological replicates. Green points indicate eCLIP-enriched peaks identified in replicate 1 (p-value ≤ 10-5 & fold-enrichment ≥ 8), with the distribution of these peaks across both replicates indicated by attached histograms.
Supplementary Figure 13 Scalable RBP target identification with eCLIP.
(a) Non-crosslinked samples show decreased RNA recovery. Bars indicate Ct value obtained by performing qPCR on 1:10 diluted pre-PCR (post-adapter ligated) library of HNRNPK and LIN28B eCLIP performed on two UV-crosslinked replicates, one non-crosslinked sample, and the paired SMInput. Increased qPCR Ct (greater required amplification) indicates decreased material obtained from the eCLIP procedure. (b) Distinct RNA binding profiles identified by eCLIP. Five HepG2 eCLIP experiments (along with paired SMInputs) are shown for the ~7kb region at the 3′ end of the RRBP1 gene, with peak calls indicated as boxes below RPM-normalized read density tracks. Significantly enriched peaks are indicated as darkened boxes. (c) Correlation between required amplification and percent of reads that are usable (i.e., not PCR duplicates) for 277 sequenced eCLIP libraries with more than 106 uniquely mapped reads. Each point indicates the eCT (extrapolated number of PCR cycles required to obtain 100 fMoles of library (x-axis), and the corresponding fraction of usable reads (out of uniquely mapped) obtained after high-throughput sequencing (y-axis). (d) eCLIP (204 libraries comprising 102 experiments in biological duplicate) yields increased usable reads with standard sequencing depth compared to 127 published iCLIP datasets or 152 published CLIP experiments. Each dataset is indicated by a point, with smoothed density plots created with the distributionPlot Matlab package with default settings (smoothened using ksdensity with a Normal kernel).
Supplementary Figure 14 eCLIP enables distinction between significant binding and common background.
(a-b) Read density tracks indicate eCLIP and SMInput signal at two abundant small RNAs. (a) A U6 snRNA (Gencode ID RNU6-9) shows read density across all eCLIP and SMInput samples, including CLIPper-called clusters (light colored bars below tracks). Significantly enriched signal is only observed for eCLIP of SMNDC1 and PRPF8, with significantly enriched peaks indicated below (as darkly colored bars). (b) Similar analysis indicates common background signal at 7SK snRNA (RN7SK), but significant enrichment in eCLIP of known 7SK ribonucleoprotein particle component LARP7. (c) Analysis of PRPF8 clusters indicates that whereas the vast majority of intronic and CDS clusters show enrichment in PRPF8 eCLIP relative to SMInput, chrM, snoRNA, and rRNA-overlapping clusters are typically false positives that are depleted in eCLIP. Notably, unlike RBFOX2 (Figure 4a), snRNA clusters identified for PRPF8 are largely enriched in PRPF8 CLIP, consistent with its known role as a core spliceosome component.
Supplementary Figure 15 RNA-centric view of RNA binding protein association.
(a-b) Sorting all 204 K562 and HepG2 eCLIP datasets by fold-enrichment for 7SK snRNA (RN7SK) identifies LARP7 as specifically binding 7SK. (a) Each bar indicates fold-enrichment in eCLIP compared to SMInput for usable reads mapping to the 7SK snRNA. (b) Bars indicate RPM of 7SK snRNA in each eCLIP dataset, before SMInput normalization. 7SK has over 100 reads in nearly all eCLIP experiments, and over 1,000 reads in the majority of experiments (datasets are ordered identically as in (a)). (c) Sorting all eCLIP experiments by fold-enrichment summed over all histone RNAs identifies SLBP as uniquely binding to histone transcripts. (d) Sorting 120 K562 eCLIP experiments by fold-enrichment for XIST identifies four proteins with greater than 2-fold enrichment: HNRNPK, PTBP1, HNRNPM, and SRSF1. (e) For the four proteins with enriched binding to XIST identified in (d), read density tracks across XIST identify specific regions of binding. Bars below density tracks indicate clusters, with significantly SMInput-enriched clusters indicated by darkened color. (f) Bars indicate RPM across lncRNA MALAT for all 204 K562 and HepG2 eCLIP datasets. (g) Tracks show read density (in RPM) across MALAT1 for eight RBPs indicated in Figure 4c, with paired SMInput datasets below. Boxes indicate CLIPper-identified clusters, with significantly enriched peaks indicated as dark boxes.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Table 3, and Supplementary Protocol 1 and 2 (PDF 15574 kb)
Supplementary Table 1
Public CLIP dataset listing and associated read mapping values. (XLSX 34 kb)
Supplementary Table 2
eCLIP experiments deposited at the ENCODE Data Coordination Center, and associated read mapping values. (XLSX 26 kb)
Source data
Rights and permissions
About this article
Cite this article
Van Nostrand, E., Pratt, G., Shishkin, A. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13, 508–514 (2016). https://doi.org/10.1038/nmeth.3810
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3810
This article is cited by
-
Dysregulation of RNA splicing in early non-alcoholic fatty liver disease through hepatocellular carcinoma
Scientific Reports (2024)
-
Towards in silico CLIP-seq: predicting protein-RNA interaction via sequence-to-signal learning
Genome Biology (2023)
-
SRSF2 is required for mRNA splicing during spermatogenesis
BMC Biology (2023)
-
A fast non-parametric test of association for multiple traits
Genome Biology (2023)
-
Comprehensive mapping of exon junction complex binding sites reveals universal EJC deposition in Drosophila
BMC Biology (2023)