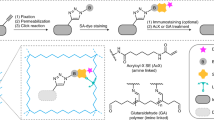
Abstract
Expansion microscopy is a technique in which fluorophores on fixed specimens are linked to a swellable polymer that is physically expanded to enable super-resolution microscopy with ordinary microscopes. We have developed and characterized new methods for linking fluorophores to the polymer that now enable expansion microscopy with conventional fluorescently labeled antibodies and fluorescent proteins. Our methods simplify the procedure and expand the palette of compatible labels, allowing rapid dissemination of the technique.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Chen, F., Tillberg, P.W. & Boyden, E.S. Science 347, 543–548 (2015).
Huang, B., Babcock, H. & Zhuang, X. Cell 143, 1047–1058 (2010).
Hell, S.W. Science 316, 1153–1158 (2007).
Hern, D.L. & Hubbell, J.A. J. Biomed. Mater. Res. 39, 266–276 (1998).
Dempsey, G.T., Vaughan, J.C., Chen, K.H., Bates, M. & Zhuang, X. Nat. Methods 8, 1027–1036 (2011).
Olivier, N., Keller, D., Gönczy, P. & Manley, S. PLoS ONE 8, e69004 (2013).
Weston, P.D. & Avrameas, S. Biochem. Biophys. Res. Commun. 45, 1574–1580 (1971).
Lee, K., Choi, S., Yang, C., Wu, H.-C. & Yu, J. Chem. Commun. (Camb.) 49, 3028–3030 (2013).
Chung, K. et al. Nature 497, 332–337 (2013).
Yang, B. et al. Cell 158, 945–958 (2014).
Migneault, I., Dartiguenave, C., Bertrand, M.J. & Waldron, K.C. BioTechniques 37, 790–802 (2004).
Kim, D. & Park, K. Polymer 45, 189–196 (2004).
Voeltz, G.K., Prinz, W.A., Shibata, Y., Rist, J.M. & Rapoport, T.A. Cell 124, 573–586 (2006).
Acknowledgements
This work is supported by the University of Washington (J.C.V.), a Burroughs-Wellcome Career Award at the Scientific Interface (J.C.V.), an NSF Graduate Research Fellowship DGE-1256082 (T.J.C.), and by NIH grants EY10699 and EY17101 (R.O.L.W.). The authors would like to thank L. Wordeman (University of Washington, Seattle, WA) for providing the PtK1 cell line and anti-HEC1 antibody, for access to an electroporator, and for helpful discussions; K. Oda (University of Washington, Seattle, WA) for performing the cardiac perfusion of mice; T. Rapoport (Harvard Medical School, Boston, MA) for the Sec61β-GFP plasmid; E. Boyden, F. Chen, and P. Tillberg (MIT, Boston, MA) for conducting an ExM workshop; and the Biology Imaging Facility at the University of Washington for imaging assistance.
Author information
Authors and Affiliations
Contributions
T.J.C., A.R.H., H.O., R.O.L.W., and J.C.V. designed the experiments. T.J.C., A.R.H., H.O., H.-J.K., and G.J.T. performed the experiments and analysis. T.J.C., A.R.H., and J.C.V. wrote the paper and all authors commented on the manuscript. J.C.V. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors have filed a patent application (U.S. Provisional 62/311, 638).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–21, Supplementary Table 1 and Supplementary Protocol (PDF 3472 kb)
Supplementary Analysis
Example data and processing scripts for pre-expansion/post-expansion distortion analysis. (ZIP 4233 kb)
3D animation of expanded brain slice shown in Fig. 3 a-f.
The animation begins as 29 × 13 μm and then zooms to show a view of 4.6 × 2.7 μm (all distances are in pre-expansion dimensions). (MOV 14725 kb)
Source data
Rights and permissions
About this article
Cite this article
Chozinski, T., Halpern, A., Okawa, H. et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods 13, 485–488 (2016). https://doi.org/10.1038/nmeth.3833
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3833
This article is cited by
-
Combined expansion and STED microscopy reveals altered fingerprints of postsynaptic nanostructure across brain regions in ASD-related SHANK3-deficiency
Molecular Psychiatry (2024)
-
Photo-expansion microscopy enables super-resolution imaging of cells embedded in 3D hydrogels
Nature Materials (2023)
-
Expanded vacuum-stable gels for multiplexed high-resolution spatial histopathology
Nature Communications (2023)
-
Imaging brain tissue architecture across millimeter to nanometer scales
Nature Biotechnology (2023)
-
Single-molecule localization microscopy reveals the ultrastructural constitution of distal appendages in expanded mammalian centrioles
Nature Communications (2023)