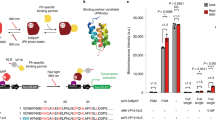
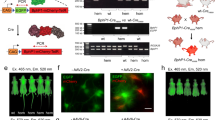
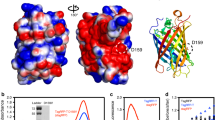
Abstract
Upon illumination, photosensitizer molecules produce reactive oxygen species that can be used for functional manipulation of living cells, including protein inactivation, targeted-damage introduction and cellular ablation. Photosensitizers used to date have been either exogenous, resulting in delivery and removal challenges, or genetically encoded proteins that form or bind a native photosensitizing molecule, resulting in a constitutively active photosensitizer inside the cell. We describe a genetically encoded fluorogen-activating protein (FAP) that binds a heavy atom−substituted fluorogenic dye, forming an 'on-demand' activated photosensitizer that produces singlet oxygen and fluorescence when activated with near-infrared light. This targeted and activated photosensitizer (TAPs) approach enables protein inactivation, targeted cell killing and rapid targeted lineage ablation in living larval and adult zebrafish. The near-infrared excitation and emission of this FAP-TAPs provides a new spectral range for photosensitizer proteins that could be useful for imaging, manipulation and cellular ablation deep within living organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lavis, L.D. & Raines, R.T. Bright ideas for chemical biology. ACS Chem. Biol. 3, 142–155 (2008).
Levskaya, A., Weiner, O.D., Lim, W.A. & Voigt, C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Lee, H.M., Larson, D.R. & Lawrence, D.S. Illuminating the chemistry of life: design, synthesis, and applications of “caged” and related photoresponsive compounds. ACS Chem. Biol. 4, 409–427 (2009).
Dolmans, D.E.J.G.J., Fukumura, D. & Jain, R.K. Photodynamic therapy for cancer. Nat. Rev. Cancer 3, 380–387 (2003).
Jacobson, K., Rajfur, Z., Vitriol, E. & Hahn, K. Chromophore-assisted laser inactivation in cell biology. Trends Cell Biol. 18, 443–450 (2008).
Allison, R.R. et al. Photosensitizers in clinical PDT. Photodiagnosis Photodyn. Ther. 1, 27–42 (2004).
Castano, A.P., Demidova, T.N. & Hamblin, M.R. Mechanisms in photodynamic therapy: part one-photosensitizers, photochemistry and cellular localization. Photodiagnosis Photodyn. Ther. 1, 279–293 (2004).
Josefsen, L.B. & Boyle, R.W. Photodynamic therapy: novel third-generation photosensitizers one step closer? Br. J. Pharmacol. 154, 1–3 (2008).
Bulina, M.E. et al. A genetically encoded photosensitizer. Nat. Biotechnol. 24, 95–99 (2006).
Shu, X. et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9, e1001041 (2011).
Westberg, M., Holmegaard, L., Pimenta, F.M., Etzerodt, M. & Ogilby, P.R. Rational design of an efficient, genetically encodable, protein-encased singlet oxygen photosensitizer. J. Am. Chem. Soc. 137, 1632–1642 (2015).
Tour, O., Meijer, R.M., Zacharias, D.A., Adams, S.R. & Tsien, R.Y. Genetically targeted chromophore-assisted light inactivation. Nat. Biotechnol. 21, 1505–1508 (2003).
Lovell, J.F., Liu, T.W.B., Chen, J. & Zheng, G. Activatable photosensitizers for imaging and therapy. Chem. Rev. 110, 2839–2857 (2010).
McDonnell, S.O. et al. Supramolecular photonic therapeutic agents. J. Am. Chem. Soc. 127, 16360–16361 (2005).
Zheng, G. et al. Photodynamic molecular beacon as an activatable photosensitizer based on protease-controlled singlet oxygen quenching and activation. Proc. Natl. Acad. Sci. USA 104, 8989–8994 (2007).
Tian, J. et al. Cell-specific and pH-activatable rubyrin-loaded nanoparticles for highly selective near-infrared photodynamic therapy against cancer. J. Am. Chem. Soc. 135, 18850–18858 (2013).
Szent-Gyorgyi, C. et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat. Biotechnol. 26, 235–240 (2008).
Saurabh, S. et al. Multiplexed modular genetic targeting of quantum dots. ACS Nano 8, 11138–11146 (2014).
Grover, A. et al. Genetically encoded pH sensor for tracking surface proteins through endocytosis. Angew. Chem. Int. Ed. Engl. 51, 4838–4842 (2012).
Saunders, M.J., Block, E., Sorkin, A., Waggoner, A.S. & Bruchez, M.P. A bifunctional converter: fluorescein quenching scfv/fluorogen activating protein for photostability and improved signal to noise in fluorescence experiments. Bioconjug. Chem. 25, 1556–1564 (2014).
Szent-Gyorgyi, C. et al. Malachite green mediates homodimerization of antibody V–L domains to form a fluorescent ternary complex with singular symmetric interfaces. J. Mol. Biol. 425, 4595–4613 (2013).
Koziar, J.C. & Cowan, D.O. Photochemical heavy-atom effects. Acc. Chem. Res. 11, 334–341 (1978).
Gandin, E., Lion, Y. & Van de Vorst, A. Quantum yield of singlet oxygen production by xanthene derivatives. Photochem. Photobiol. 37, 271–278 (1983).
Yogo, T., Urano, Y., Ishitsuka, Y., Maniwa, F. & Nagano, T. Highly efficient and photostable photosensitizer based on BODIPY chromophore. J. Am. Chem. Soc. 127, 12162–12163 (2005).
Lindig, B.A., Rodgers, M.A.J. & Schaap, A.P. Determination of the Lifetime of Singlet Oxygen in D2O Using 9,10-Anthracenedipropionic Acid, a Water-Soluble Probe. J. Am. Chem. Soc. 102, 5590–5593 (1980).
Davila, J. & Harriman, A. Photosensitized oxidation of biomaterials and related model compounds. Photochem. Photobiol. 50, 29–35 (1989).
Ogilby, P.R. Singlet oxygen: there is indeed something new under the sun. Chem. Soc. Rev. 39, 3181–3209 (2010).
Shibuya, T. & Tsujimoto, Y. Deleterious effects of mitochondrial ROS generated by KillerRed photodynamic action in human cell lines and C. elegans. J. Photochem. Photobiol. B 117, 1–12 (2012).
Qi, Y.B., Garren, E.J., Shu, X., Tsien, R.Y. & Jin, Y. Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG. Proc. Natl. Acad. Sci. USA 109, 7499–7504 (2012).
Williams, D.C. et al. Rapid and permanent neuronal inactivation in vivo via subcellular generation of reactive oxygen with the use of KillerRed. Cell Rep. 5, 553–563 (2013).
Patterson, M.S., Wilson, B.C. & Graff, R. In vivo tests of the concept of photodynamic threshold dose in normal rat-liver photosensitized by aluminum chlorosulfonated phthalocyanine. Photochem. Photobiol. 51, 343–349 (1990).
Schäfer, M. et al. Systematic study of parameters influencing the action of Rose Bengal with visible light on bacterial cells: comparison between the biological effect and singlet-oxygen production. Photochem. Photobiol. 71, 514–523 (2000).
Kuimova, M.K., Yahioglu, G. & Ogilby, P.R. Singlet oxygen in a cell: spatially dependent lifetimes and quenching rate constants. J. Am. Chem. Soc. 131, 332–340 (2009).
Tilly, J.L. & Tilly, K.I. Inhibitors of oxidative stress mimic the ability of follicle-stimulating-hormone to suppress apoptosis in cultured rat ovarian follicles. Endocrinology 136, 242–252 (1995).
Simon, H.U., Haj-Yehia, A. & Levi-Schaffer, F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 5, 415–418 (2000).
Zhao, H. et al. Superoxide reacts with hydroethidine but forms a fluorescent product that is distinctly different from ethidium: potential implications in intracellular fluorescence detection of superoxide. Free Radic. Biol. Med. 34, 1359–1368 (2003).
Fuchs, Y. & Steller, H. Programmed cell death in animal development and disease. Cell 147, 742–758 (2011).
Dodd, A., Curtis, P.M., Williams, L.C. & Love, D.A. Zebrafish: bridging the gap between development and disease. Hum. Mol. Genet. 9, 2443–2449 (2000).
Lieschke, G.J. & Currie, P.D. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367 (2007).
Curado, S., Stainier, D.Y. & Anderson, R.M. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 3, 948–954 (2008).
Poss, K.D., Wilson, L.G. & Keating, M.T. Heart regeneration in zebrafish. Science 298, 2188–2190 (2002).
Kikuchi, K. et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601–605 (2010).
Jopling, C. et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609 (2010).
Telmer, C.A. et al. Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins. ACS Chem. Biol. 10, 1239–1246 (2015).
Molina, G.A., Watkins, S.C. & Tsang, M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev. Biol. 7, 62 (2007).
Pugach, E.K., Li, P., White, R. & Zon, L. Retro-orbital injection in adult zebrafish. J. Vis. Exp. 34, 1645 (2009).
Missinato, M.A., Tobita, K., Romano, N., Carroll, J.A. & Tsang, M. Extracellular component hyaluronic acid and its receptor Hmmr are required for epicardial EMT during heart regeneration. Cardiovasc. Res. 107, 487–498 (2015).
Acknowledgements
This work was supported in part with funds from the US National Institutes of Health (NIH), Technology Centers for Networks and Pathways program (grant U54GM103529 to M.P.B., S.C.W., J.H. and Y.W.), NIH grant R01EB017268 (to J.H., Y.W., M.A.M., E.O., C.M.S., M.T. and M.P.B.) and NIH grant R21ES025606 (to M.P.B. and S.C.W.). We thank E. Kelley and C.J. Bakkenist for helpful discussion and guidance on establishing ROS involved in cellular toxicity; G. Daskivich for help in establishing zebrafish lines; and E. Drill, C.T. Wallace and M.A. Ross for help in larval zebrafish TUNEL imaging.
Author information
Authors and Affiliations
Contributions
J.H. designed and performed experiments, analyzed data and wrote the paper. Y.W. provided cell culture, performed experiments and wrote the paper. E.O., M.A.M. and M.T. developed and provided zebrafish lines, designed and performed experiments and analyzed data. L.A.P. provided reagents. S.C.W. and C.M.C. performed experiments and analyzed data. M.P.B. designed experiments, analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
M.P.B. is a founder in Sharp Edge Labs, a company applying the FAP-fluorogen technology commercially.
Integrated supplementary information
Supplementary Figure 1 Characterization of FAP-TAPs binding and spectroscopic properties.
(a) Dissociation equilibrium constant (Kd) of MG-ester–dL5** and MG-2I–dL5**, where 5 nM of protein was complexed with various concentrations of dyes. Fluorescent intensity was measured, normalized and analyzed by a one-site binding ligand depletion model. (n = 4, mean and S.E.M. plotted) (b) Fluorescent quantum yield measurement of MG-2I–dL5** using Cy5 as standard (emission spectra were integrated, normalized to the maximum value and plotted against absorbance at 600 nm).
Supplementary Figure 2 FAP-TAPs’s singlet-oxygen generation efficiency at different excitation wavelengths measured by ADPA bleaching assay.
(a) Normalized singlet oxygen yield using % ADPA bleaching by FAP-TAPs in PBS buffer with different excitation wavelengths, YFP (500/24 nm) and RFP (531/40 nm) excitation resulted in little ADPA bleaching. (n = 4, mean and S.E.M. plotted) (b) Overlap of common fluorescent proteins’ excitations with MG-2I–dL5** spectrum.
Supplementary Figure 3 ADPA bleaching in deuterated (d) PBS and photoconversion of FAP-TAPs.
(a) Percentage of ADPA bleaching by FAP-TAPs in deuterated PBS buffer; conditions are the same as in Figure 1c. (n = 4, mean and S.E.M. plotted) (b) Photoconversion of DAB by FAP-TAPs. The absorbance for each group at 669 nm was adjusted to the same. Illumination light source: LED, 669 nm, 89 mW cm-2. Clear precipitates were observed from MG-2I–dL5** and AlPcS4 after 1 hour illumination. (c) Estimated fold of activation of singlet oxygen generation from MG-2I bound to FAP, % ADPA bleaching by free MG-2I (in dPBS) (OD = 0.017, 0.041, 0.087, 0.163) at 669 nm after 60 s illumination, and by MG-2I–dL5** (in 1: 9 H2O: dPBS) (OD = 0.068, 0.148, 0.194) at 669 nm after 10 s illumination was plotted against 669 nm absorbance. Taking illumination time, bleaching rate and OD666nm difference into count, the overall fold of activation is estimated to be higher than 450-fold. (n = 4, mean and S.E.M. plotted)
Supplementary Figure 4 Representative images of MG-2I−induced cytotoxicity on HEK cells expressing FAP in the transmembrane, mitochondria, nucleus and cytosol, and of KillerRed−induced cytotoxicity targeting mitochondria.
FAP expressing cells were mixed with wild type HEK cells and then incubated with 400 nM of MG-2I (Each condition was repeated at least 4 times). Images were taken at the same conditions as for illumination (c560 of KillerRed was taken at 30% laser power to avoid saturation, 100% laser power used for illumination). Illumination conditions are denoted on the images in the ‘after illumination’ column, cell viability was determined with Live/Dead cell fluorescent assay 3 hour post illumination. (Scale bar = 10 μm)
Supplementary Figure 5 Light box (built by M.P.B.).
The box consists of a heatsink cooled 100 W deep-red LED array (LED World, 660 nm, 6000 lm specified) powered by a 24 V power supply, controlled with a CraLab enlarger timer. A cooling fan blows air through the chamber to prevent heating of the specimens during prolonged exposures. This device can also be inverted for illumination of fish, and has relatively flat-field illumination (< 5% variation) across a 96-well plate area with 0.089 W cm-2 power density (24 nm Full Width at Half Maximum). A thermometer can monitor the temperature inside the chamber to ensure samples are kept at or below physiological temperatures.
Supplementary Figure 6 Phototoxicity of TAPs on FAP-expressing HEK cells.
(a) Dose dependent cytotoxicity by MG-2I on HEK cells expressing cell surface dL5** with various illumination intensity (1 minute illumination time with 25%, 40%, 50%, 60%, 70%, 80%, 90%, 100% laser power) or illumination duration (50% laser power with 1, 2, 3, 4, 5 minute illumination time) using confocal microscopy (40× objective, 640 nm). (n = 3, mean and S.E.M. plotted) (b) Phototoxicity was suppressed only by sodium azide, but not catalase or SOD, indicating the cytotoxicity was singlet oxygen mediated. ROS quenchers were added to cells 5 minutes prior to illumination and allowed 5 minutes rescue after illumination before Live/Dead staining as above. (n = 4, mean and S.E.M. plotted)
Supplementary Figure 7 FAP-TAPs cytotoxicity rescued by various ROS quenchers.
TM -dL5** HEK cells were labeled with 400 nM of MG-2I and various ROS quenchers. After 1 minute of illumination (40× objective, 640 nm, 0.76 W cm-2), cell viability was determined with Live/Dead cell fluorescent assay. (Scale bar = 10 μm and applied to all images)
Supplementary Figure 8 Detection of superoxide species from FAP-TAPs ROS-generating cascade by DHE.
(a) Wild type HEK cells or cells expressing FAP with a nuclear localization signal (NLS-dL5** HEK) were first labeled with 400 nM of dye, and then 2 μM of DHE was added to cells 10 minutes prior to 2 minutes of illumination (60× objective, 640 nm, 2.43 W cm-2). Imaging was carried out immediately after photosensitization. (b) Mean fluorescence intensity change after illumination of each group. (n = 6, mean and S.E.M plotted. Scale bar = 10 μm and applied to all images, 488 channel: 488 nm excitation and 535/25 nm emission filter).
Supplementary Figure 9 Specific mCer3-dL5** expression and labeling, and analysis of cardiac ablation.
(a) Specific expression of Tgpt22 and labeling by MG-2I or MG-ester, images were taken by a confocal microscope, 20×, 100% laser power. Scale bar = 50 μm and applied to all images. (b) Region of interest selected to quantitatively analyze development of zebrafish from 0hpi to 96hpi. Red: MG-2I/Tgpt22, blue: MG-ester/Tgpt22, green: MG-2I/WT. In MG-2I/Tgpt22 group, the fish developed smaller eyes (****), shorter body length (*), larger edema (****) compared to MG-2I/WT and loss of mCer3 positive cell (****) compared to MG-ester/Tgpt22. Scale bar = 500 μm, n = 6, mean and S.E.M. plotted, unpaired t test, *P < 0.05, **P < 0.01; ***P < 0.001.
Supplementary Figure 10 In vivo assessment of collateral damage using larvae transgenic zebrafish; apoptosis was restricted to FAP-TAPs−targeted cells.
Fish were fixed 24 hpi and stained using a whole mount TUNEL assay and immunolabeling of the mCer3 with anti-GFP antibodies and a secondary alexa555 conjugated donkey anti-rabbit antibody. (a) Representative imaging of orthgonal view and sphere reconstruction of TUNEL/mCer3 fluorescence. (b) mCer3+ cell count, fewer mCer3+ cells are seen in MG-2I 12 min group (*). (c) TUNEL positive cell count, green represents TUNEL signals that colocalized with mCer3 signals; black represents TUNEL signals that were not associated with mCer3+ cells. (NS for all groups) (d) TUNEL/mCer3+ ratio, MG-2I with 12 min illumination induced 31.5% (**) cell death of mCer3+ cells compared to MG-ester with 12 min group. A reduced fraction of mCer3+ cells are killed (24.8%) when 4 min illumination is applied (*). Scale Bar = 10 µm and applied to all images, n = 8, mean and S.E.M plotted. One-way ANOVA, Tukey post hoc tests were performed with multiple comparisons of mean for each group. Paired t test was used for comparisons between specific death and non-specific death. *P < 0.05, **P < 0.01.
Supplementary Figure 11 FAP-TAPs−induced photo-ablation is specific to the cardiac tissue in adult zebrafish.
(a) Tgpt23 zebrafish uniformly express mCer3-dL5** at the adult stage in the atrium (A) and in the ventricle (V). (b) Schematic illustration of experimental setup for photo-ablation of adult zebrafish. (c) TUNEL assay in liver and intestine (Gut) at 3 dpi showed no differences between the four groups. (d) AFOG staining of liver and Gut at 5 dpi showing no structural damage in all groups. n = 5 for each group, One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001; scale bar = 100 µm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1 and 2, and Supplementary Note (PDF 2242 kb)
Change in cell morphology after photosensitization.
The first frame is an image of FAP-TAPs fluorescence (640 channel) before illumination; the last frame was an image of EthD-1 fluorescence (560 channel, dead cell stain) and taken 30 minutes after illumination. Scale bar, 10 μm. (MP4 1212 kb)
Fluorescence bleaching of FAP-TAPs on cell surface with 10 min of continuous illumination using a confocal microscope (40×, 640 nm, 0.43 W/cm2).
Less than 30% of self-bleaching was observed in the first one minute of illumination, which was the typical duration for inducing sufficient cytotoxicity. Scale bar, 10 μm. (MP4 243 kb)
A Tg(myl7:MBIC5-mCer3) larvae labeled with 500 nM MG-ester at 72 h.p.f. (incubated for 3 h).
Movies were taken with a confocal microscope, 20×, 640 nm, 100% laser power (640 channel in red). (MP4 309 kb)
Tg(myl7:MBIC5-mCer3) larvae treated with different conditions: MG-2I and 20-min illumination, MG-2I and 5-min illumination, MG-2I and no light, and MG-ester and 20-min illumination.
Movies were taken right after illumination using a Canon camera (10×, 470/40 ex, 510/40 em, green). (MP4 697 kb)
Source data
Rights and permissions
About this article
Cite this article
He, J., Wang, Y., Missinato, M. et al. A genetically targetable near-infrared photosensitizer. Nat Methods 13, 263–268 (2016). https://doi.org/10.1038/nmeth.3735
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3735
This article is cited by
-
PARP2 promotes Break Induced Replication-mediated telomere fragility in response to replication stress
Nature Communications (2024)
-
Genetically targeted chemical assembly
Nature Reviews Bioengineering (2023)
-
Global and transcription-coupled repair of 8-oxoG is initiated by nucleotide excision repair proteins
Nature Communications (2022)
-
Telomeric 8-oxo-guanine drives rapid premature senescence in the absence of telomere shortening
Nature Structural & Molecular Biology (2022)
-
Functions of ADP-ribose transferases in the maintenance of telomere integrity
Cellular and Molecular Life Sciences (2022)