Abstract
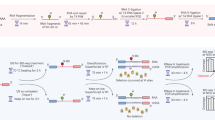
Despite its biological importance, tRNA has not been adequately sequenced by standard methods because of its abundant post-transcriptional modifications and stable structure, which interfere with cDNA synthesis. We achieved efficient and quantitative tRNA sequencing in HEK293T cells by using engineered demethylases to remove base methylations and a highly processive thermostable group II intron reverse transcriptase to overcome these obstacles. Our method, DM-tRNA-seq, should be applicable to investigations of tRNA in all organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wang, Z., Gerstein, M. & Snyder, M. Nat. Rev. Genet. 10, 57–63 (2009).
Pang, Y.L., Abo, R., Levine, S.S. & Dedon, P.C. Nucleic Acids Res. 42, e170 (2014).
Kirchner, S. & Ignatova, Z. Nat. Rev. Genet. 16, 98–112 (2015).
Abbott, J.A., Francklyn, C.S. & Robey-Bond, S.M. Front. Genet. 5, 158 (2014).
Trewick, S.C., Henshaw, T.F., Hausinger, R.P., Lindahl, T. & Sedgwick, B. Nature 419, 174–178 (2002).
Falnes, P.Ø., Johansen, R.F. & Seeberg, E. Nature 419, 178–182 (2002).
Zheng, G., Fu, Y. & He, C. Chem. Rev. 114, 4602–4620 (2014).
Katibah, G.E. et al. Proc. Natl. Acad. Sci. USA 111, 12025–12030 (2014).
Shen, P.S. et al. Science 347, 75–78 (2015).
Mohr, S. et al. RNA 19, 958–970 (2013).
Goodenbour, J.M. & Pan, T. Nucleic Acids Res. 34, 6137–6146 (2006).
Chan, P.P. & Lowe, T.M. Nucleic Acids Res. 37, D93–D97 (2009).
Dittmar, K.A., Goodenbour, J.M. & Pan, T. PLoS Genet. 2, e221 (2006).
Gingold, H. et al. Cell 158, 1281–1292 (2014).
Pavon-Eternod, M. et al. Nucleic Acids Res. 37, 7268–7280 (2009).
Horton, R. et al. Nat. Rev. Genet. 5, 889–899 (2004).
Holland, P.J. & Hollis, T. PLoS ONE 5, e8680 (2010).
Yu, B. et al. Nature 439, 879–884 (2006).
Mishina, Y., Chen, L.X. & He, C. J. Am. Chem. Soc. 126, 16930–16936 (2004).
Zheng, G. et al. Mol. Cell 49, 18–29 (2013).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grant DP1GM105386 to T.P., NIH grants R01GM37949 and GM37951 to A.M.L., K01HG006699 to Q.D., NIH MCB Training Grant (T32GM007183) to W.C.C. and a Chicago Biomedical Consortium Postdoctoral Research Grant Award to G.Z. We thank L. Zhang, W.J. Chen and I.A. Gagnon for technical assistance. C.H. is supported by Howard Hughes Medical Institute as an investigator.
Author information
Authors and Affiliations
Contributions
G.Z., Y.Q., W.C.C., Q.D., A.M.L. and T.P. designed and performed experiments and analyzed data. G.Z. and T.P. conceived the project. G.Z., C.Y. and C.H. designed the demethylase constructs. G.Z., Y.Q., W.C.C., A.M.L. and T.P. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
Thermostable group II intron reverse transcriptase (TGIRT) enzymes and methods for their use are the subject of patents and patent applications that have been licensed by the University of Texas at Austin and East Tennessee State University to InGex, LLC. A.M.L. and the University of Texas are minority equity holders in InGex, LLC, and A.M.L. and other present and former members of the Lambowitz laboratory receive royalty payments from sales of TGIRT enzymes and licensing of intellectual property.
Integrated supplementary information
Supplementary Figure 1 Development of DM-tRNA-seq.
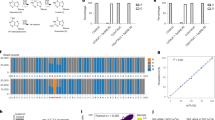
(a) View of AlkB active site stereochemistry models with m1G coordination based on Protein data bank (PDB) ID 3KHC and 3BIE. Bottom left shows m1G bound wtAlkB while bottom right shows m1G bound by the D135S mutant. The mutated amino acid is indicated in red. (b) Demethylation efficiency of proteins towards modifications. tRNAs were treated with equimolar AlkB (pink), equimolar D135S (cyan), or a mix of both AlkB and D135S (tRNA: AlkB: D135S = 1:1:1) at pH 5. Demethylation fractions of m1A (top) and m1G (bottom) were subsequently analyzed. (c) The pH-activity profiles for demethylation reactions of m1A and m1G in tRNA by a mix of wtAlkB and D135S. (d) The protein ratio profiles for demethylation reactions of m1A and m1G. tRNA was incubated with equimolar wtAlkB with varying folds of D135S as indicated in the figure, since wtAlkB has already been shown to efficiently demethylate m1A. All the demethylation experiments shown were carried out in triplicate; error bar, n = 3 ± SD.
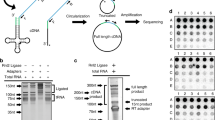
Supplementary Figure 2 DM-tRNA-seq mapping.
(a) Total reads, mapped reads, and mapped rate for 4 sets of biological replicates. (b) Compare added internal tRNA standards and total reads; error bar, n = 4 ± SD. Despite the wide variations of total reads in each sample, reads of the standards are very similar, indicating that the read variations among individual samples do not reflect the variations in tRNA abundance, rather, they were derived from variations in sample handling and efficiency of enzyme reactions in each sample.
Supplementary Figure 3 Replicate sequencing plots.
(a) Purified tRNA, untreated. (b) Purified tRNA, +demethylases. (c) Total RNA, untreated. (d) Total RNA, +demethylases.
Supplementary Figure 4 Chromosome 6 tRNA isodecoder expression.
Among all human chromosomes, Chr. 6 contains the highest number of annotated tRNA genes (166), plus 9 tRNA pseudogenes. Of these 175 genes/pseudogenes, 157 are clustered within a ~2.7 Mbp region next to the class I MHC genes. We were able to determine the expression of 131 genes/pseudogenes because of their unique sequences among themselves or among other Chr.6 tRNA genes. The other 44 genes have the same sequence as one or more tRNA genes located in other chromosomes. (a) Untreated. Arrow points to the expanded region containing the tRNA gene cluster within the 2.7 Mbp region. The number of tRNA genes that can be uniquely analyzed is indicated: 117 in the 2.7 Mbp region, and 14 outside this region. (b) Plus demethylase treatment.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 341 kb)
Source data
Rights and permissions
About this article
Cite this article
Zheng, G., Qin, Y., Clark, W. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12, 835–837 (2015). https://doi.org/10.1038/nmeth.3478
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3478
This article is cited by
-
tRNA therapeutics for genetic diseases
Nature Reviews Drug Discovery (2024)
-
Roles and regulation of tRNA-derived small RNAs in animals
Nature Reviews Molecular Cell Biology (2024)
-
Small RNA structural biochemistry in a post-sequencing era
Nature Protocols (2024)
-
Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing
Nature Biotechnology (2024)
-
The role and mechanism of action of tRNA-derived fragments in the diagnosis and treatment of malignant tumors
Cell Communication and Signaling (2023)