Abstract
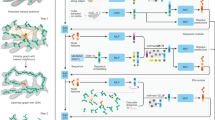
We present a de novo model-building approach that combines predicted backbone conformations with side-chain fit to density to accurately assign sequence into density maps. This method yielded accurate models for six of nine experimental maps at 3.3- to 4.8-Å resolution and produced a nearly complete model for an unsolved map containing a 660-residue heterodimeric protein. This method should enable rapid and reliable protein structure determination from near-atomic-resolution cryo-electron microscopy (cryo-EM) maps.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hryc, C.F., Chen, D.H. & Chiu, W. Curr. Opin. Virol. 1, 110–117 (2011).
Liao, M., Cao, E., Julius, D. & Cheng, Y. Nature 504, 107–112 (2013).
Bai, X.C., Fernandez, I.S., McMullan, G. & Scheres, S.H. eLife 2, e00461 (2013).
Lu, P. et al. Nature 512, 166–170 (2014).
Rossmann, M.G., Bernal, R. & Pletnev, S.V. J. Struct. Biol. 136, 190–200 (2001).
DiMaio, F. et al. Nat. Methods doi:10.1038/nmeth.3286 (23 February 2015).
Allegretti, M., Mills, D.J., McMullan, G., Kühlbrandt, W. & Vonck, J. eLife 3, e01963 (2014).
Amunts, A. et al. Science 343, 1485–1489 (2014).
Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006).
Terwilliger, T.C. et al. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
Baker, M.R., Rees, I., Ludtke, S.J., Chiu, W. & Baker, M.L. Structure 20, 450–463 (2012).
Lindert, S. et al. Structure 17, 990–1003 (2009).
Gront, D., Kulp, D.W., Vernon, R.M., Strauss, C.E. & Baker, D. PLoS ONE 6, e23294 (2011).
Song, Y. et al. Structure 21, 1735–1742 (2013).
Kudryashev, M. et al. Cell 10.1016/j.cell.2015.01.037 (in the press).
Bradley, P., Misura, K.M. & Baker, D. Science 309, 1868–1871 (2005).
Raman, S. et al. Science 327, 1014–1018 (2010).
DiMaio, F.P., Soni, A.B., Phillips, G.N. Jr. & Shavlik, J.W. Int. J. Data Min. Bioinform. 3, 205–227 (2009).
DiMaio, F., Tyka, M.D., Baker, M.L., Chiu, W. & Baker, D. J. Mol. Biol. 392, 181–190 (2009).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Winn, M.D. et al. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Chen, X., Wang, Q., Ni, F. & Ma, J. Proc. Natl. Acad. Sci. USA 107, 11352–11357 (2010).
Mindell, J.A. & Grigorieff, N. J. Struct. Biol. 142, 334–347 (2003).
Roseman, A.M. J. Struct. Biol. 145, 91–99 (2004).
Frank, J. et al. J. Struct. Biol. 116, 190–199 (1996).
Li, X., Grigorieff, N. & Cheng, Y. J. Struct. Biol. 172, 407–412 (2010).
Li, X. et al. Nat. Methods 10, 584–590 (2013).
Scheres, S.H. & Chen, S. Nat. Methods 9, 853–854 (2012).
Rosenthal, P.B. & Henderson, R. J. Mol. Biol. 333, 721–745 (2003).
Acknowledgements
The authors thank K. Laidig and D. Alonso for setting up and managing the computational resources. This work was supported by the US National Institutes of Health under award numbers R01GM092802 (R.Y.-R.W. and D.B.), EB001567 (E.H.E.) and R01GM098672 (Y.C.); the Swiss systems biology initiative SystemsX.ch grant CINA (M.K.); the University of California, San Francisco, Program for Breakthrough Biomedical Research (Y.C.); and Howard Hughes Medical Institute (D.B.). We thank Y. Liu for the use of his clustering algorithm.
Author information
Authors and Affiliations
Contributions
R.Y.-R.W. performed the research and drafted the manuscript. F.D. and D.B. supervised the research and edited the draft. M.K., E.H.E. and M.B. provided the helical reconstruction map of the VipA/VipB complex and compared the automated model with their manually traced model initially. X.L. and Y.C. provided the 20S map at 4.8-Å resolution. All authors edited the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 RosettaCM fixes placement errors in the final partial model of FrhA.
(a) One case where there is an error in fragment placement. (b) Density-guided rebuilding in RosettaCM is able to correct this error as the missing residues are rebuilt. The backbone trace in grey indicates the native structure (4ci0).
Supplementary Figure 2 Problems in building β-sheets.
Each row indicates a case where our automatic approach failed to determine the structure: VP6 at 3.8 Å (top) and STIV at 3.9 Å (bottom). Illustrated are the density map (left), the partial model at the iteration with highest accuracy (middle), and the native structure (right).
Supplementary Figure 3 Real-space density correlation over ambiguous region in the map.
A structural alignment of the residues with a discrepancy in sequence registration is indicated on the x-axis (the residues in between the orange and blue arrows in Figure 3a and 3b). The sequence assignment of the automated and manual models is labeled on the upper and lower axes, respectively. The blue line (automated model) and the red line (manual model) show the real-space density correlation for each assignment (y-axis). Real-space density correlation was calculated using density_tools in Rosetta with a 2.5Å mask.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Tables 1–3 (PDF 1340 kb)
Rights and permissions
About this article
Cite this article
Wang, RR., Kudryashev, M., Li, X. et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat Methods 12, 335–338 (2015). https://doi.org/10.1038/nmeth.3287
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3287
This article is cited by
-
CR-I-TASSER: assemble protein structures from cryo-EM density maps using deep convolutional neural networks
Nature Methods (2022)
-
Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14
Nature Communications (2021)
-
Structure of the p53/RNA polymerase II assembly
Communications Biology (2021)
-
Deep Learning to Predict Protein Backbone Structure from High-Resolution Cryo-EM Density Maps
Scientific Reports (2020)
-
Macromolecular modeling and design in Rosetta: recent methods and frameworks
Nature Methods (2020)