Abstract
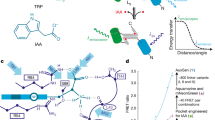
The visualization of hormonal signaling input and output is key to understanding how multicellular development is regulated. The plant signaling molecule auxin triggers many growth and developmental responses, but current tools lack the sensitivity or precision to visualize these. We developed a set of fluorescent reporters that allow sensitive and semiquantitative readout of auxin responses at cellular resolution in Arabidopsis thaliana. These generic tools are suitable for any transformable plant species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
24 September 2014
In the version of this article initially published, it was stated that all transgenic lines were created in the Arabidopsis Col-0 ecotype. However, they were actually generated in the Columbia-Utrecht (Col-utr) ecotype. The error has been corrected in the HTML and PDF versions of the article.
References
Lokerse, A.S. & Weijers, D. Curr. Opin. Plant Biol. 12, 520–526 (2009).
Muday, G.K. J. Plant Growth Regul. 20, 226–243 (2001).
Dharmasiri, N., Dharmasiri, S. & Estelle, M. Nature 435, 441–445 (2005).
Kepinski, S. & Leyser, O. Nature 435, 446–451 (2005).
Gray, W.M., Kepinski, S., Rouse, D., Leyser, O. & Estelle, M. Nature 414, 271–276 (2001).
Tan, X. et al. Nature 446, 640–645 (2007).
Ulmasov, T., Hagen, G. & Guilfoyle, T.J. Science 276, 1865–1868 (1997).
Ulmasov, T., Hagen, G. & Guilfoyle, T.J. Proc. Natl. Acad. Sci. USA 96, 5844–5849 (1999).
Wang, R. & Estelle, M. Curr. Opin. Plant Biol. 21, 51–58 (2014).
Ulmasov, T., Murfett, J., Hagen, G. & Guilfoyle, T.J. Plant Cell 9, 1963–1971 (1997).
Friml, J. et al. Nature 426, 147–153 (2003).
Moreno-Risueno, M.A. et al. Science 329, 1306–1311 (2010).
Jones, A.R. et al. Nat. Cell Biol. 11, 78–84 (2009).
Scarpella, E., Marcos, D., Friml, J. & Berleth, T. Genes Dev. 20, 1015–1027 (2006).
Grieneisen, V.A., Xu, J., Marée, A.F.M., Hogeweg, P. & Scheres, B. Nature 449, 1008–1013 (2007).
Boer, D.R. et al. Cell 156, 577–589 (2014).
Hardtke, C.S. et al. Development 131, 1089–1100 (2004).
Perrot-Rechenmann, C. Cold Spring Harb. Perspect. Biol. 2, a001446 (2010).
Brunoud, G. et al. Nature 482, 103–106 (2012).
Völker, A., Stierhof, Y.D. & Jürgens, G. J. Cell Sci. 114, 3001–3012 (2001).
Weijers, D. et al. Development 128, 4289–4299 (2001).
Federici, F., Dupuy, L., Laplaze, L., Heisler, M. & Haseloff, J. Nat. Methods 9, 483–485 (2012).
Wend, S. et al. Sci. Rep. 3, 2052 (2013).
Robert, H.S. et al. Curr. Biol. 23, 2506–2512 (2013).
Vernoux, T. et al. Mol. Syst. Biol. 7, 508 (2011).
De Rybel, B.D. et al. Plant Physiol. 156, 1292–1299 (2011).
Willemsen, V. et al. Plant Cell 15, 612–625 (Humana Press, 2003).
Llavata-Peris, C., Lokerse, A., Möller, B., De Rybel, B. & Weijers, D. in Plant Organogenesis: Methods and Protocols (ed. De Smet, Ive) Ch. 8, 137–148 (Humana Press, 2013).
van den Berg, C., Willemsen, V., Hage, W., Weisbeek, P. & Scheres, B. Nature 378, 62–65 (1995).
Daghma, D.S., Kumlehn, J., Hensel, G., Rutten, T. & Melzer, M. J. Exp. Bot. 63, 6017–6021 (2012).
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F. & Vandesompele, J. Genome Biol. 8, R19 (2007).
Acknowledgements
We thank T. Laux (Universität Freiburg) for plasmids and B. de Rybel for helpful comments on the manuscript. This work was supported by grants from the European Research Council (ERC; CELLPATTERN; contract number 281573) and the Netherlands Organization for Scientific Research (NWO; ALW-820.02.019) to D.W. and Human Frontier Science Program (HFSP; research grant RGP0054-2013) and Agence Nationale de la Recherche (ANR; AuxiFlo; grant ANR-12-BSV6-0005) to T.V.
Author information
Authors and Affiliations
Contributions
C.-Y.L. generated all transgenic lines with the exception of RPS5A-DII-Venus lines, which were generated by G.B. All imaging was performed by C.-Y.L. and W.S. S.Y. contributed to analysis of DII-Venus lines. D.W. and T.V. supervised the project. C.-Y.L. and D.W. conceived of the study and wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
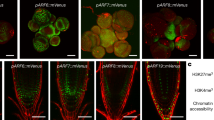
Supplementary Figure 1 Overview comparison of DR5 and DR5v2 activity in root tip.
Maximal projection of propidium iodine stained root of (a) DR5:: n3eGFP and (b) DR5v2:: n3eGFP reporter lines. Scale bars are 10 µm.
Supplementary Figure 2 Performance of primers against tandem repeated reporter genes.
qRT-PCR of serial diluted serial diluted pGIIM/DR5v2::ntdTomato-DR5::n3eGFP plasmid with primers used in this study. Bars indicate standard error from the mean (n=3).
Supplementary Figure 3 Response of DR5 and DR5v2 to external auxin.
Fluorescent signal intensity of n3xGFP (top row) and ntdTomato (bottom row) in DR5v2::ntdTomato-DR5::n3eGFP root tips following a 12-hour co-treatment of 10 µM NPA and the indicated concentrations of IAA. Detector gain was saturated for each channel separately at the highest signal intensity of the 1000 nM IAA treated root, and all other images were acquired using these same settings. Signal intensity is displayed as a false color scale. Scale bars are 10 µm.
Supplementary Figure 4 pRPS5a::DII:Venus and pRPS5a::mDII:Venus root tips.
(a) pRPS5a:: DII: Venus and (b) pRPS5a:: mDII: Venus. Scale bars are 10 µm.
Supplementary Figure 5 DR5v2-n3GFP and R2D2 heart-stage embryos.
Scale bars are 10 µm.
Supplementary Figure 6 Quantification of R2D2 gradients.
Normalized ntdTomato/n3xVenus signal ratio in nuclei at increasing distance from the QC (see dashed lines in image on the right). Cell 1 corresponds to the first daughter of the initial for each cell file. Red/yellow ratio was set to “1” in cell 1 for each cell file. Bars indicate standard error from the mean (n>30 cell files per tissue). Scale bars are 10 µm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1 and Supplementary Note 1 (PDF 1731 kb)
R2D2 activity upon auxin treatment
Overlay of red and green fluorescence in a root after treatment with 1 μM IAA during 33 minutes (1 frame every 3 minutes). (AVI 8450 kb)
R2D2 activity upon control treatment
Overlay of red and green fluorescence in a root after treatment with control medium during 33 minutes (1 frame every 3 minutes). (AVI 8450 kb)
Rights and permissions
About this article
Cite this article
Liao, CY., Smet, W., Brunoud, G. et al. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12, 207–210 (2015). https://doi.org/10.1038/nmeth.3279
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3279
This article is cited by
-
Two orthogonal differentiation gradients locally coordinate fruit morphogenesis
Nature Communications (2024)
-
Highly efficient Agrobacterium rhizogenes-mediated transformation for functional analysis in woodland strawberry
Plant Methods (2023)
-
Antigravitropic PIN polarization maintains non-vertical growth in lateral roots
Nature Plants (2023)
-
Epidermal injury-induced derepression of key regulator ATML1 in newly exposed cells elicits epidermis regeneration
Nature Communications (2023)
-
Seeing is understanding
Nature Plants (2023)