Abstract
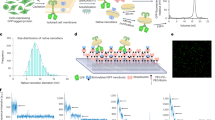
Proteoliposome reconstitution is a standard method to stabilize purified transmembrane proteins in membranes for structural and functional assays. Here we quantified intrareconstitution heterogeneities in single proteoliposomes using fluorescence microscopy. Our results suggest that compositional heterogeneities can severely skew ensemble-average proteoliposome measurements but also enable ultraminiaturized high-content screens. We took advantage of this screening capability to map the oligomerization energy of the β2-adrenergic receptor using ∼109-fold less protein than conventional assays.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rigaud, J.L. & Lévy, D. in Methods in Enzymology: Liposomes, Part B Vol. 372 (ed. Duzgunes, N.) 65–86 (Elsevier, 2003).
Seddon, A.M., Curnow, P. & Booth, P.J. Biochim. Biophys. Acta 1666, 105–117 (2004).
Serebryany, E., Zhu, G.A. & Yan, E.C.Y. Biochim. Biophys. Acta 1818, 225–233 (2012).
Niu, S.L., Doctrow, B. & Mitchell, D.C. Biochemistry 48, 156–163 (2009).
Mansoor, S.E., Palczewski, K. & Farrens, D.L. Proc. Natl. Acad. Sci. USA 103, 3060–3065 (2006).
Larsen, J., Hatzakis, N.S. & Stamou, D. J. Am. Chem. Soc. 133, 10685–10687 (2011).
Chen, X. et al. Biophys. J. 90, 2062–2074 (2006).
Kimura, T. et al. J. Biol. Chem. 287, 4076–4087 (2012).
Bomholt, J. et al. Langmuir 27, 866–869 (2011).
Cowell, G.M., Tranum-Jensen, J., Sjöström, H. & Norén, O. Biochem. J. 237, 455–461 (1986).
Diao, J. et al. Nat. Protoc. 7, 921–934 (2012).
Grasso, L. et al. PLoS ONE 8, e70929 (2013).
Christensen, A.L., Lohr, C., Christensen, S.M. & Stamou, D. Lab Chip 13, 3613–3625 (2013).
Bendix, P.M., Pedersen, M.S. & Stamou, D. Proc. Natl. Acad. Sci. USA 106, 12341–12346 (2009).
Stamou, D., Duschl, C., Delamarche, E. & Vogel, H. Angew. Chem. Int. Ed. Engl. 42, 5580–5583 (2003).
Audet, M. & Bouvier, M. Cell 151, 14–23 (2012).
Overington, J.P., Al-Lazikani, B. & Hopkins, A.L. Nat. Rev. Drug Discov. 5, 993–996 (2006).
Milligan, G. Mol. Pharmacol. 84, 158–169 (2013).
Ciruela, F., Vilardaga, J.P. & Fernández-Dueñas, V. Trends Biotechnol. 28, 407–415 (2010).
Wolber, P.K. & Hudson, B.S. Biophys. J. 28, 197–210 (1979).
Adair, B.D. & Engelman, D.M. Biochemistry 33, 5539–5544 (1994).
Yano, Y. & Matsuzaki, K. Biochemistry 45, 3370–3378 (2006).
Leslie, M. Science 331, 24–26 (2011).
Caruso, F. Colloids and Colloid Assemblies: Synthesis, Modification, Organization and Utilization of Colloid Particles (Wiley, 2006).
Fung, J.J. et al. EMBO J. 28, 3315–3328 (2009).
Rasmussen, S.G.F. et al. Nature 450, 383–387 (2007).
Xie, G., Gross, A.K. & Oprian, D.D. Biochemistry 42, 1995–2001 (2003).
Ridge, K.D., Lu, Z., Liu, X. & Khorana, H.G. Biochemistry 34, 3261–3267 (1995).
Fay, J.F. & Farrens, D.L. J. Biol. Chem. 287, 33873–33882 (2012).
Tsukamoto, H., Sinha, A., DeWitt, M. & Farrens, D.L. J. Mol. Biol. 399, 501–511 (2010).
Ghanouni, P. et al. J. Biol. Chem. 276, 24433–24436 (2001).
Fay, J.F., Dunham, T.D. & Farrens, D.L. Biochemistry 44, 8757–8769 (2005).
Periasamy, A., Wallrabe, H., Chen, Y. & Barroso, M. Methods Cell Biol. 89, 569–598 (2008).
McCann, J.J., Choi, U.B., Zheng, L.Q., Weninger, K. & Bowen, M.E. Biophys. J. 99, 961–970 (2010).
Lakowicz, J.R. Principles of Fluorescence Spectroscopy 3rd edn. (Springer, 2006).
Magde, D., Brannon, J.H., Cremers, T.L. & Olmsted, J. J. Phys. Chem. 83, 696–699 (1979).
Sauer, M. et al. J. Fluoresc. 5, 247–261 (1995).
Kunding, A.H., Mortensen, M.W., Christensen, S.M. & Stamou, D. Biophys. J. 95, 1176–1188 (2008).
Ulbrich, M.H. & Isacoff, E.Y. Nat. Methods 4, 319–321 (2007).
Marrink, S.J., de Vries, A.H. & Mark, A.E. J. Phys. Chem. B 108, 750–760 (2004).
Acknowledgements
This work was supported by the Lundbeck Foundation, the Danish Councils for Independent and Strategic Research and the University of Copenhagen programs of excellence “Single molecule Nanoscience,” “BioScaRT” and “UNIK-Synthetic Biology.” We thank E.M. Danielsen and H. Flyvbjerg for helpful discussion.
Author information
Authors and Affiliations
Contributions
D.S. conceived of the strategy and was responsible for the overall project management and supervision. D.S., S.M.C. and S.M. designed all experiments. Fluorescence measurements, data analysis and theoretical calculations were performed by S.M. S.V. assisted with ligand binding experiments. J.J.F. and S.G.F.R. expressed, purified, labeled and reconstituted β2-AR; J.F.F. expressed, purified, labeled and reconstituted CB1 and opsin. S.K.J. and S.M.C. developed analysis algorithms and wrote Igor scripts. M.K. supported theoretical fitting and calculations. D.S., S.M. and S.M.C. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 GPCR labeling positions.
Positions of fluorescently labeled cysteines (marked by red) in the protein constructs employed in this study. β2AR pdb code 2RH1. Currently no crystal structure is available for the CB1, we therefore used a homology model based on pdb code 3V2Y, for illustrating the labeling position. Opsin pdb code 3CAP.
Supplementary Figure 2 Representative single-molecule bleaching steps for quantification of protein densities.
Single molecule bleaching steps was used as a calibration to determine the actual number of receptors on single proteoliposomes (see online Methods ). The most probable number of receptors was ∼20 for the nominal P/L 1:1,000 shown in Fig. 2c. (a) A histogram of Cy3 single molecule bleaching step fitted with a gaussian. The peak position (x0) of 2007 ± 28 is used for further analysis. (b) Example of Cy3 bleaching step. (c) A histogram of Cy5 single molecule bleaching step intensities fitted with a gaussian. The peak position (x0) of 3318 ± 117 is used for further analysis. (d) Example of Cy5 bleaching step.
Supplementary Figure 3 Distributions of number of proteins for three of the different reconstitutions shown in Figure 2b (nominal P/L = 1:50,000, 1:80,000 or 1:300,000).
(a-i) The number of proteins was determined by single molecule photobleaching (online Methods ) for each individual reconstitution. (a) Distribution of number of proteins for nominal P/L 1:50,000. (b) Empty liposomes are included in the distribution of number of proteins for nominal P/L 1:50,000. In the case that insertion of receptors into the proteoliposomes is entirely random, the number distribution of receptors should be poissonian. A mean proteoliposome diameter of 100 nm corresponds to ∼100,000 lipids pr. proteoliposome (using lipid headgroup area of 0.67 nm2). For a reconstitution of 1:50,000 nominal P/L a poissonian distribution should therefore have a mean of 2 proteins pr. liposome. This distribution is plotted in red. (c) Zoom of b. (d) Distributions of number of proteins for nominal P/L 1:80,000. (e) Empty liposomes are included in the distribution of number of proteins for nominal P/L 1:80,000. (red) poissonian distribution for a mean of 1.25 proteins pr. liposome (assuming a mean number of lipids of 100,000 pr. proteoliposome). (f) Zoom of e. (h) Distributions of number of proteins for nominal P/L 1:300,000. (i) Empty liposomes are included in the distribution of number of proteins for P/L nominal 1:300,000. (red) poissonian distribution for a mean of 0.33 proteins pr. liposome (assuming a mean number of lipids of 100,000). (j) Zoom of i.
Supplementary Figure 4 Distribution of proteoliposome diameters and receptor density versus proteoliposome diameter, as found for β2-AR (Δ5-R333C, nominal P/L = 1:1,000).
(a) Histogram of proteoliposome diameters. (b) Receptor density plotted against proteoliposome diameter.
Supplementary Figure 5 Quantification of FRET correction factors.
(a) Cy3 (green) and Cy5 (red) absorption and emission spectra is shown together with donor excitation (543 nm, black vertical line) and acceptor emission bandpass filter (blue). Orange marks the direct excitation of acceptor by donor excitation 543 nm (α). Light green area marks the donor intensity in Cy5 detection channel β. (b) Histogram of the percentage donor intensity in acceptor channel at donor excitation (exc. 543 nm), β. β was calculated as the ratio of donor signal arising in the acceptor channel (exc. 543 nm) to the donor signal in the donor channel (exc. 543 nm). β was determined for 1,130 single proteoliposomes carrying only β2AR-Cy3 (nominal P/L 1:1,000). (c) Histogram of percentage direct excitation of acceptor at donor excitation (exc. 543 nm), α. α was calculated as the ratio of acceptor signal (exc. 543 nm) in the acceptor channel to the signal of the acceptor excited at 633 nm. α was determined for 729 single proteoliposomes carrying only β2AR-Cy5 (nominal P/L 1:1,000). (d) Histogram of the percentage Oregon Green-DHPE in donor channel at donor excitation (exc. 543 nm), ω. ω was calculated as the ratio of Oregon Green-DHPE intensity (exc. 543 nm) in the donor channel to Oregon Green-DHPE excited at 476 nm. ω was determined for 729 single liposomes carrying only Oregon Green-DHPE. (e) Histogram of single proteoliposome EFRET (blue) and corresponding histogram of EFRET errors (red) (β2AR nominal P/L 1:1,000). The calculated ensemble average FRET efficiency EFRET,bulk is marked by the dark gray line.
Supplementary Figure 6 Comparing the measured FRET efficiencies with FRET arising from random collisions.
(a) The predicted FRET efficiency of random collisions (colorscale) as accessed by eq. 7 in Supplementary Note for the range of reduced acceptor densities Cα and reduced total den. Ct measured in a single reconstitution sample (β2AR, P/L 1:1,000). (b) For comparison the measured EFRET (colorscale). The measured EFRET is well above random collisions, and supports that we observe significant oligomerization.
Supplementary Figure 7 The effect of ligands on β2-AR oligomerization.
(a-c) Reduced total receptor density Ct and reduced acceptor density Ca as a function of EFRET (color scale) for a population of proteoliposomes with a narrow size distribution of 120-130 nm. For better visualization EFRET values of single liposomes in a-c are binned and a weighed average of each bin is shown. (a) No ligand treatment, N = 921 proteoliposomes. (b) Proteoliposomes incubated with saturating amounts (10 μM) of agonist Isoproterenol (ISO), N = 321 proteoliposomes. (c) Proteoliposomes incubated with saturating amounts (500 nM) of inverse agonist ICI 118,551 (ICI). N = 510 proteoliposomes.
Supplementary Figure 8 GPCR size-exclusion chromatography elution profiles.
(a) Size exclusion chromatography (SEC) trace of detergent solubilized β2AR-365 functionally purified by sequential antibody and ligand affinity chromatography. (b) Full-length β2AR was functionally purified in absence of reducing reagent tris(2-carboxyethyl)phosphine (TCEP) and iodoacetamide (IA) allowing cysteine bridge formation between receptors. Size exclusion chromatography of the purified β2AR was performed in absence (red) and following (black) of TCEP/IA pretreatment. In reducing conditions the fraction of monomeric detergent solubilized receptors increase and the dimeric/oligomeric species decrease. (c-d) Fluorescence-detected size-exclusion chromatography (FSEC) traces of purified Cy5 labeled detergent solubilized CB1 and Opsin. About 25 pmoles of sample was injected into a 100 uL loading loop and then run over a hand packed 60 mL (34cm x 1.5 cm diameter) Superdex 200 (prep grade) column using 20 mM TRIS pH 7.5, 150 mM NaCl, 1mM EDTA and 0.05% DM at a flow rate of 0.5 mL/min. The elutions were monitored via a RF-551 fluorescence HPLC monitor (Shimadzu) with excitation wavelength set at 635 nm and emission wavelength set at 665 nm for Cy5. Elution time correspond to ∼85 kDa for opsin and ∼175 kDa for CB1 as estimated from the their molecular weights (38.5 kDa and ∼40 kDa) and the addition of a DM micelle. The markers: 1, 2, 3, & 4 represent molecular weight standards (void volume, 669 kDa, 43 kDa, and 27 kDa, respectively).
Supplementary Figure 9 Theoretical determination of (ηA/ηD).
(a) Transmission efficiency (left) for Cy3 bandpass filter (HC585/40), Cy5 filter HQ675/55M, and beamsplitter BS625. Detection efficiency for the APD (right) of type SPCM-AQR-13-FCM, Perkin Elmer. (b) Initial and final Cy3 and Cy5 emission spectra. Normalized Cy3 and Cy5 emission spectra and the reduced final emission after passing through each element in the optical pathway 1) beamsplitter 2) filter 3) APD, shown in a.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Note and Supplementary Discussion (PDF 1438 kb)
Supplementary Software
Single liposome FRET analysis software developed in Igor Pro ver. 6.01. (ZIP 20 kb)
Rights and permissions
About this article
Cite this article
Mathiasen, S., Christensen, S., Fung, J. et al. Nanoscale high-content analysis using compositional heterogeneities of single proteoliposomes. Nat Methods 11, 931–934 (2014). https://doi.org/10.1038/nmeth.3062
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3062
This article is cited by
-
Regulation of the mammalian-brain V-ATPase through ultraslow mode-switching
Nature (2022)
-
Using single-vesicle technologies to unravel the heterogeneity of extracellular vesicles
Nature Protocols (2021)
-
Permanent Photodynamic Cholecystokinin 1 Receptor Activation: Dimer-to-Monomer Conversion
Cellular and Molecular Neurobiology (2018)
-
Membrane curvature regulates ligand-specific membrane sorting of GPCRs in living cells
Nature Chemical Biology (2017)
-
Dynamics of P-type ATPase transport revealed by single-molecule FRET
Nature (2017)