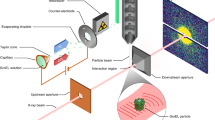
Abstract
We describe a method to measure ultrafast protein structural changes using time-resolved wide-angle X-ray scattering at an X-ray free-electron laser. We demonstrated this approach using multiphoton excitation of the Blastochloris viridis photosynthetic reaction center, observing an ultrafast global conformational change that arises within picoseconds and precedes the propagation of heat through the protein. This provides direct structural evidence for a 'protein quake': the hypothesis that proteins rapidly dissipate energy through quake-like structural motions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Ansari, A. et al. Proc. Natl. Acad. Sci. USA 82, 5000–5004 (1985).
Miyashita, O., Onuchic, J.N. & Wolynes, P.G. Proc. Natl. Acad. Sci. USA 100, 12570–12575 (2003).
Vos, M.H., Rappaport, F., Lambry, J.-C., Breton, J. & Martin, J.-L. Nature 363, 320–325 (1993).
Rischel, C. et al. Proc. Natl. Acad. Sci. USA 95, 12306–12311 (1998).
Wang, H. et al. Science 316, 747–750 (2007).
Wang, H. et al. J. Phys. Chem. B 113, 818–824 (2009).
Cammarata, M. et al. Nat. Methods 5, 881–886 (2008).
Andersson, M. et al. Structure 17, 1265–1275 (2009).
Ahn, S., Kim, K.H., Kim, Y., Kim, J. & Ihee, H. J. Phys. Chem. B 113, 13131–13133 (2009).
Cho, H.S. et al. Proc. Natl. Acad. Sci. USA 107, 7281–7286 (2010).
Malmerberg, E. et al. Biophys. J. 101, 1345–1353 (2011).
Ramachandran, P.L. et al. J. Am. Chem. Soc. 133, 9395–9404 (2011).
Kim, K.H. et al. J. Am. Chem. Soc. 134, 7001–7008 (2012).
Takala, H. et al. Nature 509, 245–248 (2014).
Emma, P. et al. Nat. Photonics 4, 641–647 (2010).
Boutet, S. & Williams, G.W. New J. Phys. 12, 035024 (2010).
Weierstall, U., Spence, J.C. & Doak, R.B. Rev. Sci. Instrum. 83, 035108 (2012).
Palazzo, G., Lopez, F. & Mallardi, A. Biochim. Biophys. Acta 1804, 137–146 (2010).
Go, N., Noguti, T. & Nishikawa, T. Proc. Natl. Acad. Sci. USA 80, 3696–3700 (1983).
Cook, D.J. & Hochstrasser, R.M. Opt. Lett. 25, 1210–1212 (2000).
DePonte, D.P. et al. J. Phys. D Appl. Phys. 41, 195505 (2008).
Hart, P. et al. Proc. SPIE 8504, 85040C (2012).
Georgiou, P. et al. J. Chem. Phys. 124, 234507 (2006).
Clayton, R.K. & Clayton, B.J. Biochim. Biophys. Acta 501, 478–487 (1978).
Valkunas, L. & Gulbinas, V. Photochem. Photobiol. 66, 628–634 (1997).
Valkunas, L., Trinkunas, G., Liuolia, V. & van Grondelle, R. Biophys. J. 69, 1117–1129 (1995).
Kasha, M. Discuss. Faraday Soc. 9, 14–19 (1950).
Cammarata, M. et al. Rev. Sci. Instrum. 80, 015101 (2009).
Deisenhofer, J., Epp, O., Sinning, I. & Michel, H. J. Mol. Biol. 246, 429–457 (1995).
Duan, Y. et al. J. Comput. Chem. 24, 1999–2012 (2003).
Ceccarelli, M., Procacci, P. & Marchi, M. J. Comput. Chem. 24, 129–142 (2003).
Autenrieth, F., Tajkhorshid, E., Baudry, J. & Luthey-Schulten, Z. J. Comput. Chem. 25, 1613–1622 (2004).
Gast, P., Hemelrijk, P. & Hoff, A.J. FEBS Lett. 337, 39–42 (1994).
Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A. & Haak, J.R. J. Chem. Phys. 81, 3684–3690 (1984).
Bussi, G., Donadio, D. & Parrinello, M. J. Chem. Phys. 126, 014101 (2007).
Hess, B., Bekker, H., Berendsen, H.J.C. & Fraaije, J.G.E.M. J. Comput. Chem. 18, 1463–1472 (1997).
Essmann, U. et al. J. Chem. Phys. 103, 8577–8593 (1995).
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. J. Chem. Theory Comput. 4, 435–447 (2008).
Breton, J., Martin, J.L., Migus, A., Antonetti, A. & Orszag, A. Proc. Natl. Acad. Sci. USA 83, 5121–5125 (1986).
Acknowledgements
Experiments were carried out at the LCLS, a national user facility operated by Stanford University on behalf of the US Department of Energy (DOE), Office of Basic Energy Sciences. We acknowledge financial support from the Swedish Science Research Council (VR), the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), the Swedish Strategic Research Foundation (SSF), the Knut and Alice Wallenberg Foundation, the US National Science Foundation (NSF) and its bioXFEL Science and Technology Center (NSF 1231306), the US National Institute of Health (NIH), the DOE Office of Basic Energy Sciences, the Hamburg Ministry of Science and Research, the Joachim Herz Stiftung, the Deutsche Forschungsgemeinschaft (DFG), the German Federal Ministry for Education and Research (BMBF), a Marie Curie International Incoming Fellowship of the European Union, the Academy of Finland, the Max Planck Society, the Danish National Research Foundations Centre for Molecular Movies, DANSCATT, the UCOP Lab Fee Program (award no. 118036) and the LLNL Lab-directed Research & Development Program (12-ERD-031).
Author information
Authors and Affiliations
Contributions
R.N. and D.A. conceived of the experiment, which was designed with E.M., J.D., A.B., H.N.C., J.C.H.S., S. Boutet, G.J.W., D.P.D., R.B.D. and U.W. Samples were prepared by D.A. and L.C.J. Time-resolved WAXS experiments at the LCLS were performed by D.A., L.C.J., A.B., G.J.W., C.W., E.M., J.D., D.M., D.P.D., R.L.S., D.W., D.J., G.K., S.W., T.A.W., A.A., S. Bari, M.B., R.B.D., M.F., R.F., I.G., M.S.H., R.A.K., C.K., M.L., A.V.M., M.M., M.M.S., J.S., F.S., U.W., N.A.Z., P.F., I.S., S. Boutet, H.N.C. and R.N. Time-resolved WAXS experiments at Argonne National Laboratory were performed by D.A., L.C.J., P.B., J.S., J.D., R.H. and I.K. The CXI instrument was set up by S. Boutet, G.J.W., M.M. and M.M.S., and the on-line femtosecond laser was set up by D.M., J.D. and M.F. Samples were delivered by R.L.S., S. Bari, D.P.D., U.W. and R.B.D. Data were analyzed by D.A., A.B., E.M., C.W. and R.N. MD simulations of RCvir were performed by G.G., theoretical fitting was performed by D.A., C.W. and R.N., and heat flow dynamics were examined by G.G., K.S.K., T.B.v.D. and M.M.N. The manuscript was prepared by R.N., D.A. and G.G. with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Structure of the B. viridis photosynthetic reaction center.
RCvir consists of transmembrane subunits L and M that support a special pair of bacteriochlorophylls (P960), two monomeric bacteriochlorophylls (BCh), two bacteriopheophytins molecules (BPh), a tightly bound menaquinone (QA) and a mobile quinone (QB) arranged around an axis of two-fold pseudo-symmetry. RCvir is capped from the cytoplasm by the H-subunit and from the periplasm by the tetraheme containing C-subunit. Photo-oxidation of P960 causes an electron to move to QB on the opposite side of the membrane (red arrow) whereupon P960+ is reduced from the heme groups of the C-subunit. A second photo-oxidation reaction causes the QB- semiquinone to be fully reduced to quinol, protonated from the cytoplasm and released into the membrane. Blue lines indicate approximate membrane boundaries. In this work multi-photon excitation was used, resulting in the rapid heating of the BCh cofactors and the single-photon electron transfer scheme above does not necessarily apply.
Supplementary Figure 2 Absorption spectra from detergent-solubilized RCvir before and after exposure to a single 800-nm femotsecond laser pulse.
RCvir samples were exposed to a pump laser of 500 fs in duration tuned to a fluence of 4.4 mJ mm-2 per pulse and operated at a repetition rate of 1 kHz. Samples were passed through a 300 μm quartz capillary and diluted to achieve OD800 nm = 1 through this capillary. The capillary was placed in the focus of the fs laser beam. The rate of flow of the sample through the quartz capillary was chosen so that each and every sample volume was exposed once, and only once, to the 800 nm pulse. Samples were collected and steady-state spectra (shown) were taking after the sample was loaded into a 10 mm path length cuvette and diluted 200 fold. The red line shows absorption spectra recorded from samples exposed to the fs laser, whereas the black line corresponds to samples that were not exposed. These spectra are averaged over three repeats.
Supplementary Figure 3 Changes in WAXS difference spectra due to structural changes in membrane proteins.
Comparison of the protein difference WAXS basis spectrum for RCvir (C2 of Fig. 1d, blue) with two basis spectra recorded using TR-WAXS studies of bacteriorhodopsin (bR): magenta, the basis spectrum arising from bR with a time-constant of 22 μs; green the basis spectrum arising from bR with a time-constant of 1.9 ms; blue, the basis spectrum arising from RCvir with a half rise of 1.4 ps. The two bR basis spectra have been arbitrarily scaled to assist comparison with the RCvir basis spectrum.
Supplementary Figure 4 Changes in WAXS spectra from detergent-solubilized RCvir due to heating.
(a) Temperature dependence of ΔS(q) measured by continuously heating the sample when held in a capillary at a synchrotron radiation source. The inset (0.2 Å-1 ≤ q ≤ 0.8 Å-1) illustrates that the oscillatory features observed in ultrafast time-resolved WAXS data (Fig. 1) do not arise when continuously heating the sample. The temperature changes used for these measurements were 5 °C (blue; also inset); 10 °C (green); 15 °C (magenta) and 20 °C (light blue). (b) Measurements of ΔS(q,Δt) recorded using 100 ps X-ray pulses of synchrotron radiation for the time delays Δt = 280 ps and Δt = 1 μs. (c) Overlay of the ΔS(q, Δt = 280 ps) from XFEL data (green) with ΔS(q) corresponding to a 7 °C temperature change (blue). The blue curve was recovered by interpolating the data in a and simulating random fluctuations in the background associated with XFEL data (Online Methods).
Supplementary Figure 5 Time-dependent linear amplitudes of the WAXS basis spectra extracted by spectral decomposition.
(a) Time dependence of the ultrafast (C1) component. (b) Time dependence of the protein (C2) component. (c) Time dependence of the non-equilibrated heating (C3) component. (d) Time dependence of the equilibrated heating (C4) component. Linear amplitudes of these components are shown as blue stars and the time dependent amplitudes used for spectral decomposition are shown as continuous lines. Colors chosen to be consistent with Fig. 1.
Supplementary Figure 6 Recurring movements extracted from an ensemble-average approach to structural fitting against the experimental C2 basis spectrum.
Bottom left: Matrix representation of the average internal distance changes calculated on Cα atoms, Avg([ΔCα]ij), divided by the standard deviation of these changes, [σ(ΔCα)]ij. This matrix was calculated from an ensemble of 720 best fits identified by χ2 minimization against the protein difference WAXS component C2. Top right: Recurring movements, defined as Avg([ΔCα]ij) ≥ 0.9 × [σ(ΔCα)]ij relative to at least 20 other Cα atoms (Online Methods). Only changes corresponding to movements of Cα atoms away from each other ([ΔCα]ij > 0) were classified as recurring, with no inwards movements ([ΔCα]ij < 0) noted above this threshold. All of these movements are associated with the TM helices of subunits H, L and M and are represented as orange spheres on the atomic coordinates in Fig. 3C.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1202 kb)
Animation of the structural information presented in Fig. 3c.
This video illustrates reoccurring light induced movements in RCvir extracted using ensemble average fitting against the TR-WAXS basis spectrum C2 (Fig. 3b). Orange spheres represent Cα atoms that move away from at least 20 other Cα atoms. These Cα atoms all lie within the TM domain of RCvir and therefore correspond to a light-induced expansion of the protein's TM domain. (MPG 18683 kb)
Animation of the light induced global conformational change in RCvir.
This video illustrates movements extracted using ensemble average fitting against the TR-WAXS basis spectrum C2 (Fig. 3b). This movie was created by morphing between structures calculated by averaging over an ensemble of photo-excited and resting state structures selected by pair-wise χ2 fitting against the C2 basis spectrum (Fig. 3b). These movements have been exaggerated four fold to better illustrate the nature of the average motion. Although this animation summarizes the structural model derived from an ensemble analysis, it should not be taken to represent a unique structural model for the conformation adopted by RCvir a few picoseconds after photoactivation. It is possible that other low-resolution models, not sampled within the photo-activated MD trajectories, could yield similar agreement to the experimental difference WAXS data. (MPG 308 kb)
Rights and permissions
About this article
Cite this article
Arnlund, D., Johansson, L., Wickstrand, C. et al. Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nat Methods 11, 923–926 (2014). https://doi.org/10.1038/nmeth.3067
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3067
This article is cited by
-
Ultrafast structural changes direct the first molecular events of vision
Nature (2023)
-
Determining the charge distribution and the direction of bond cleavage with femtosecond anisotropic x-ray liquidography
Nature Communications (2022)
-
Ultrafast coherent motion and helix rearrangement of homodimeric hemoglobin visualized with femtosecond X-ray solution scattering
Nature Communications (2021)
-
Filming ultrafast roaming-mediated isomerization of bismuth triiodide in solution
Nature Communications (2021)
-
Tracking Membrane Protein Dynamics in Real Time
The Journal of Membrane Biology (2021)