Abstract
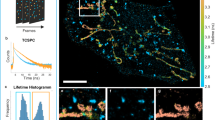
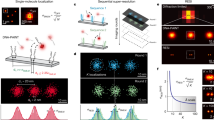
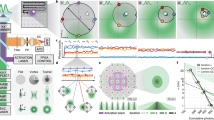
Super-resolution fluorescence microscopy is a powerful tool for biological research, but obtaining multiplexed images for a large number of distinct target species remains challenging. Here we use the transient binding of short fluorescently labeled oligonucleotides (DNA-PAINT, a variation of point accumulation for imaging in nanoscale topography) for simple and easy-to-implement multiplexed super-resolution imaging that achieves sub-10-nm spatial resolution in vitro on synthetic DNA structures. We also report a multiplexing approach (Exchange-PAINT) that allows sequential imaging of multiple targets using only a single dye and a single laser source. We experimentally demonstrate ten-color super-resolution imaging in vitro on synthetic DNA structures as well as four-color two-dimensional (2D) imaging and three-color 3D imaging of proteins in fixed cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Hell, S.W. Microscopy and its focal switch. Nat. Methods 6, 24–32 (2009).
Hell, S.W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Sharonov, A. & Hochstrasser, R.M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA 103, 18911–18916 (2006).
Giannone, G. et al. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J. 99, 1303–1310 (2010).
Lew, M.D. et al. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 108, E1102–E1110 (2011).
Flors, C., Ravarani, C.N. & Dryden, D.T. Super-resolution imaging of DNA labelled with intercalating dyes. ChemPhysChem 10, 2201–2204 (2009).
Schoen, I., Ries, J., Klotzsch, E., Ewers, H. & Vogel, V. Binding-activated localization microscopy of DNA structures. Nano Lett. 11, 4008–4011 (2011).
Jungmann, R. et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10, 4756–4761 (2010).
Tokunaga, M., Imamoto, N. & Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008).
Jungmann, R., Scheible, M. & Simmel, F.C. Nanoscale imaging in DNA nanotechnology. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 4, 66–81 (2012).
Lin, C. et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 4, 832–839 (2012).
Derr, N.D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012).
Johnson-Buck, A. et al. Super-resolution fingerprinting detects chemical reactions and idiosyncrasies of single DNA pegboards. Nano Lett. 13, 728–733 (2013).
Xu, K., Babcock, H.P. & Zhuang, X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods 9, 185–188 (2012).
Vaughan, J.C., Jia, S. & Zhuang, X. Ultrabright photoactivatable fluorophores created by reductive caging. Nat. Methods 9, 1181–1184 (2012).
Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S.M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Steinhauer, C., Jungmann, R., Sobey, T.L., Simmel, F.C. & Tinnefeld, P. DNA origami as a nanoscopic ruler for super-resolution microscopy. Angew. Chem. Int. Ed. Engl. 48, 8870–8873 (2009).
Aitken, C.E., Marshall, R.A. & Puglisi, J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008).
Rasnik, I., McKinney, S.A. & Ha, T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 3, 891–893 (2006).
Vogelsang, J. et al. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew. Chem. Int. Ed. Engl. 47, 5465–5469 (2008).
Ries, J., Kaplan, C., Platonova, E., Eghlidi, H. & Ewers, H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012).
Wu, N. et al. Molecular threading and tunable molecular recognition on DNA origami nanostructures. J. Am. Chem. Soc. 135, 12172–12175 (2013).
Kao, H.P. & Verkman, A.S. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys. J. 67, 1291–1300 (1994).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Lubeck, E. & Cai, L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods 9, 743–748 (2012).
Bates, M., Dempsey, G.T., Chen, K.H. & Zhuang, X. Multicolor super-resolution fluorescence imaging via multi-parameter fluorophore detection. ChemPhysChem 13, 99–107 (2012).
Schweller, R.M. et al. Multiplexed in situ immunofluorescence using dynamic DNA complexes. Angew. Chem. Int. Ed. Engl. 51, 9292–9296 (2012).
Ke, R. et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013).
Opazo, F. et al. Aptamers as potential tools for super-resolution microscopy. Nat. Methods 9, 938–939 (2012).
Kazane, S.A. et al. Site-specific DNA-antibody conjugates for specific and sensitive immuno-PCR. Proc. Natl. Acad. Sci. USA 109, 3731–3736 (2012).
Beliveau, B.J. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 109, 21301–21306 (2012).
Acknowledgements
We thank J. Nicoludis and M.T. Strauss for help with DNA origami design, T. Schlichthaerle for transmission electron microscopy imaging support and M. Zhang for help with DLD1 cells. We thank C. Steinhauer for help with DNA-PAINT software development and fruitful discussions. We thank R.D. Barish for critical reading and commenting on the manuscript. This work is supported by a US National Institutes of Health (NIH) Director's New Innovator Award (1DP2OD007292), an NIH Transformative Research Award (1R01EB018659), an NIH grant (5R21HD072481), an Office of Naval Research (ONR) Young Investigator Program Award (N000141110914), ONR grants (N000141010827 and N000141310593), a US National Science Foundation (NSF) Faculty Early Career Development Award (CCF1054898), an NSF grant (CCF1162459) and a Wyss Institute for Biologically Engineering Faculty Startup Fund to P.Y., and an NIH Director's New Innovator Award (1DP2OD004641) and a Wyss Institute for Biologically Inspired Engineering Faculty Award to W.M.S. R.J. acknowledges support from the Alexander von Humboldt-Foundation through a Feodor-Lynen Fellowship. M.S.A. and M.D. acknowledge support from Howard Hughes Medical Institute International Student Research Fellowships.
Author information
Authors and Affiliations
Contributions
R.J., M.S.A. and J.B.W. contributed equally to this work. R.J. and M.S.A. conceived of the study, designed and performed the experiments, analyzed the data and wrote the manuscript. J.B.W. designed and performed the experiments, analyzed the data and wrote the manuscript. M.D. performed the experiments, analyzed the data and developed the drift correction software. W.M.S. supervised the project, discussed the results and critiqued the paper. P.Y. conceived of, designed and supervised the study, interpreted the data and wrote the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.J., M.D., M.S.A., J.B.W. and P.Y. have filed a provisional US patent application regarding the current work.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Tables 1–8 and Supplementary Protocol (PDF 29243 kb)
Supplementary Software
Parallelized spot finding and 2D Gaussian fitting software implemented in LabVIEW (ZIP 1769 kb)
Source data
Rights and permissions
About this article
Cite this article
Jungmann, R., Avendaño, M., Woehrstein, J. et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods 11, 313–318 (2014). https://doi.org/10.1038/nmeth.2835
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2835
This article is cited by
-
Near-infrared PAINT localization microscopy via chromophore replenishment of phytochrome-derived fluorescent tag
Communications Biology (2024)
-
Universal, label-free, single-molecule visualization of DNA origami nanodevices across biological samples using origamiFISH
Nature Nanotechnology (2024)
-
Thermal-plex: fluidic-free, rapid sequential multiplexed imaging with DNA-encoded thermal channels
Nature Methods (2024)
-
DNA-barcoded signal amplification for imaging mass cytometry enables sensitive and highly multiplexed tissue imaging
Nature Methods (2023)
-
Single-molecule analysis of DNA base-stacking energetics using patterned DNA nanostructures
Nature Nanotechnology (2023)