Abstract
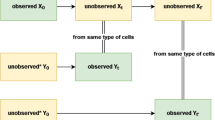
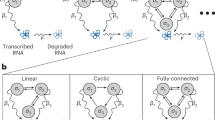
Mathematical methods combined with measurements of single-cell dynamics provide a means to reconstruct intracellular processes that are only partly or indirectly accessible experimentally. To obtain reliable reconstructions, the pooling of measurements from several cells of a clonal population is mandatory. However, cell-to-cell variability originating from diverse sources poses computational challenges for such process reconstruction. We introduce a scalable Bayesian inference framework that properly accounts for population heterogeneity. The method allows inference of inaccessible molecular states and kinetic parameters; computation of Bayes factors for model selection; and dissection of intrinsic, extrinsic and technical noise. We show how additional single-cell readouts such as morphological features can be included in the analysis. We use the method to reconstruct the expression dynamics of a gene under an inducible promoter in yeast from time-lapse microscopy data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Zechner, C. et al. Moment-based inference predicts bimodality in transient gene expression. Proc. Natl. Acad. Sci. USA 109, 8340–8345 (2012).
Hasenauer, J. et al. Identification of models of heterogeneous cell populations from population snapshot data. BMC Bioinformatics 12, 125 (2011).
Ornatsky, O. et al. Highly multiparametric analysis by mass cytometry. J. Immunol. Methods 361, 1–20 (2010).
Raj, A., Peskin, C.S., Tranchina, D., Vargas, D.Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006).
Neuert, G. et al. Systematic identification of signal-activated stochastic gene regulation. Science 339, 584–587 (2013).
Mettetal, J.T., Muzzey, D., Gómez-Uribe, C. & van Oudenaarden, A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319, 482–484 (2008).
Harper, C.V. et al. Dynamic analysis of stochastic transcription cycles. PLoS Biol. 9, e1000607 (2011).
Suter, D.M. et al. Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474 (2011).
Amrein, M. & Künsch, H.R. Rate estimation in partially observed Markov jump processes with measurement errors. Stat. Comput. 22, 513–526 (2012).
Golightly, A. & Wilkinson, D.J. Bayesian parameter inference for stochastic biochemical network models using particle Markov chain Monte Carlo. Interface Focus 1, 807–820 (2011).
Opper, M. & Sanguinetti, G. Variational inference for Markov jump processes. in Adv. Neural Inf. Process. Syst. Vol. 20 (eds. Platt, J.C., Koller, D., Singer, Y. & Roweis, D.) (MIT Press, 2009).
Stathopoulos, V. & Girolami, M.A. Markov chain Monte Carlo inference for Markov jump processes via the linear noise approximation. Philos. Trans. A Math. Phys. Eng. Sci. 371, 20110541 (2013).
Elowitz, M.B., Levine, A.J., Siggia, E.D. & Swain, P.S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Colman-Lerner, A. et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature 437, 699–706 (2005).
Snijder, B. & Pelkmans, L. Origins of regulated cell-to-cell variability. Nat. Rev. Mol. Cell Biol. 12, 119–125 (2011).
Bowsher, C.G. & Swain, P.S. Identifying sources of variation and the flow of information in biochemical networks. Proc. Natl. Acad. Sci. USA 109, E1320–E1328 (2012).
Hilfinger, A. & Paulsson, J. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proc. Natl. Acad. Sci. USA 108, 12167–12172 (2011).
Koeppl, H., Zechner, C., Ganguly, A., Pelet, S. & Peter, M. Accounting for extrinsic variability in the estimation of stochastic rate constants. Int. J. Robust Nonlinear Control 22, 1103–1119 (2012).
Aalen, O. Nonparametric inference for a family of counting processes. Ann. Stat. 6, 701–726 (1978).
Doucet, A., Freitas, N., Murphy, K. & Russell, S. Rao-Blackwellised particle filtering for dynamic Bayesian networks. in 16th Annu. Conf. Uncertain. Artif. Intell. (eds. Boutilier, C. & Godszmidt, M.) 176–183 (Morgan Kaufmann, 2000).
Rinott, R., Jaimovich, A. & Friedman, N. Exploring transcription regulation through cell-to-cell variability. Proc. Natl. Acad. Sci. USA 108, 6329–6334 (2011).
Louvion, J.F., Havaux-Copf, B. & Picard, D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene 131, 129–134 (1993).
McIsaac, R.S. et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 22, 4447–4459 (2011).
Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564 (1988).
Varshavsky, A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93, 12142–12149 (1996).
Hackett, E.A., Esch, K.R., Maleri, S. & Errede, B. A family of destabilized cyan fluorescent proteins as transcriptional reporters in S. cerevisiae. Yeast 23, 333–349 (2006).
Pelet, S., Dechant, R., Lee, S.S., van Drogen, F. & Peter, M. An integrated image analysis platform to quantify signal transduction in single cells. Integr. Biol. (Camb.) 4, 1274–1282 (2012).
Blake, W.J., Kaern, M., Cantor, C.R. & Collins, J.J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003).
Zenklusen, D., Larson, D.R. & Singer, R.H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008).
Mason, P.B. & Struhl, K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17, 831–840 (2005).
Raser, J.M. & O'Shea, E.K. Control of stochasticity in eukaryotic gene expression. Science 304, 1811–1814 (2004).
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using μManager. Curr. Prot. Mol. Biol. 92, 14.20 (2010).
Friedman, N., Cai, L. & Xie, X.S. Linking stochastic dynamics to population distribution: An analytical framework of gene expression. Phys. Rev. Lett. 97, 168302 (2006).
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010).
Wilkinson, D.J. Stochastic Modelling for Systems Biology 1st edn. (Chapman and Hall/CRC, 2006).
Küchler, U. & Sorensen, M. Exponential Families of Stochastic Processes (Springer, 1997).
Anderson, D.F. A modified next reaction method for simulating chemical systems with time dependent propensities and delays. J. Chem. Phys. 127, 214107 (2007).
Storvik, G. Particle filters for state-space models with the presence of unknown static parameters. IEEE Trans. Signal Process. 50, 281–289 (2002).
Koller, D. & Friedman, N. Probabilistic Graphical Models: Principles and Techniques (MIT Press, 2009).
Acknowledgements
We want to thank H.R. Kuensch and J. Hasenauer for their valuable feedback on the manuscript and O. Aalen for providing us with his technical report from 1988. We thank F. Rudolf for help in designing and cloning the Y-Venus destabilized reporter and S. Lee with the fluidic setup. C.Z., M.U. and H.K. acknowledge support from the Swiss National Science Foundation, grant no. PP00P2_128503 and SystemsX.ch. S.P. and M.P. acknowledge support from the European project UNICELLSYS, European Research Council, SystemsX.ch organization (LiverX), Swiss National Science Foundation and ETH Zurich. M.U. receives support from the Life Science Zurich PhD Program on Systems Biology of Complex Diseases; and M.U., M.P. and H.K. acknowledge support from the Competence Center for Systems Physiology and Metabolic Diseases, Zurich, Switzerland.
Author information
Authors and Affiliations
Contributions
C.Z., M.U., M.P. and H.K. designed the research; C.Z. and H.K. conceived of mathematical methods, performed simulations and analyzed data; M.U. and S.P. developed strains; M.U. and S.P. performed experiments and measured data; and C.Z. and H.K. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Notes 1–9 (PDF 2380 kb)
Rights and permissions
About this article
Cite this article
Zechner, C., Unger, M., Pelet, S. et al. Scalable inference of heterogeneous reaction kinetics from pooled single-cell recordings. Nat Methods 11, 197–202 (2014). https://doi.org/10.1038/nmeth.2794
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2794
This article is cited by
-
Gene expression model inference from snapshot RNA data using Bayesian non-parametrics
Nature Computational Science (2023)
-
Bye bye, linearity, bye: quantification of the mean for linear CRNs in a random environment
Journal of Mathematical Biology (2023)
-
Memory and relatedness of transcriptional activity in mammalian cell lineages
Nature Communications (2019)
-
Estimating numbers of intracellular molecules through analysing fluctuations in photobleaching
Scientific Reports (2019)
-
Multi-experiment nonlinear mixed effect modeling of single-cell translation kinetics after transfection
npj Systems Biology and Applications (2018)