Abstract
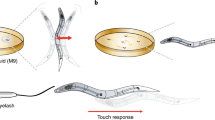
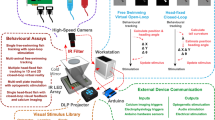
Using low-cost automated tracking microscopes, we have generated a behavioral database for 305 Caenorhabditis elegans strains, including 76 mutants with no previously described phenotype. The growing database currently consists of 9,203 short videos segmented to extract behavior and morphology features, and these videos and feature data are available online for further analysis. The database also includes summary statistics for 702 measures with statistical comparisons to wild-type controls so that phenotypes can be identified and understood by users.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ramani, A.K. et al. Cell 148, 792–802 (2012).
Houle, D., Govindaraju, D.R. & Omholt, S. Nat. Rev. Genet. 11, 855–866 (2010).
Husson, S.J., Costa, W.S., Schmitt, C. & Gottschalk, A. in WormBook (ed. Hobert, O.), 10.1895/wormbook.1.156.1 (2012).
Hart, A.C. in WormBook (ed. Ambros, V.), 10.1895/wormbook.1.87.1 (2006).
Geng, W., Cosman, P., Berry, C.C., Feng, Z. & Schafer, W.R. IEEE Trans. Biomed. Eng. 51, 1811–1820 (2004).
Cronin, C.J. et al. BMC Genet. 6, 5 (2005).
Huang, K.M., Cosman, P. & Schafer, W.R. J. Neurosci. Methods 171, 153–164 (2008).
Stephens, G.J., Johnson-Kerner, B., Bialek, W. & Ryu, W.S. PLOS Comput. Biol. 4, e1000028 (2008).
Huang, K.M., Cosman, P. & Schafer, W.R. J. Neurosci. Methods 158, 323–336 (2006).
Storey, J.D. J. R. Stat. Soc. Series B Stat. Methodol. 64, 479–498 (2002).
Starich, T.A., Xu, J., Skerrett, I.M., Nicholson, B.J. & Shaw, J.E. Neural Dev. 4, 16 (2009).
Yeh, E. et al. PLoS Biol. 6, e55 (2008).
Boulin, T. et al. Proc. Natl. Acad. Sci. USA 105, 18590–18595 (2008).
Kindt, K.S. et al. Nat. Neurosci. 10, 568–577 (2007).
Jose, A.M., Bany, I.A., Chase, D.L. & Koelle, M.R. Genetics 175, 93–105 (2007).
Feng, Z. et al. Cell 127, 621–633 (2006).
Ghosh, R., Mohammadi, A., Kruglyak, L. & Ryu, W.S. BMC Biol. 10, 85 (2012).
Brown, A.E., Yemini, E.I., Grundy, L.J., Jucikas, T. & Schafer, W.R. Proc. Natl. Acad. Sci. USA 110, 791–796 (2013).
Yemini, E., Kerr, R.A. & Schafer, W.R. Cold Spring Harb. Protoc. 2011, 1475–1479 (2011).
Tsai, C.A. & Chen, J.J. Bioinformatics 25, 897–903 (2009).
Zhao, B., Khare, P., Feldman, L. & Dent, J.A. J. Neurosci. 23, 5319–5328 (2003).
McGrath, P.T. et al. Neuron 61, 692–699 (2009).
Weber, K.P. et al. PLoS ONE 5, e13922 (2010).
Brenner, S. Genetics 77, 71–94 (1974).
Otsu, N. IEEE Trans. Syst. Man. Cyber. 9, 62–66 (1979).
Freeman, H. IRE Trans. Electron. Comput. EC-10, 260–268 (1961).
Sulston, J.E. & Horvitz, H.R. Dev. Biol. 56, 110–156 (1977).
White, J.G., Southgate, E., Thomson, J.N. & Brenner, S. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986).
Alkema, M.J., Hunter-Ensor, M., Ringstad, N. & Horvitz, H.R. Neuron 46, 247–260 (2005).
Acknowledgements
We thank C. Cronin and P. Sternberg for providing the code from their single-worm tracker, which serves as the basis for elements of our WT2 analysis GUI and phenotypic features cited from their publication; J. Lasenby and N. Kingsbury for guidance in computer vision; A. Deonarine for useful discussions; and R. Samworth, S. Chavali, G. Chalancon, S. Teichmann and M. Babu for guidance in statistics and bioinformatics techniques. This project was funded by grants to W.R.S. from the MRC (MC-A022-5PB91) and the National Institute on Drug Abuse, US National Institutes of Health (DA018341), a Gates Cambridge Scholarship (to E.Y.) and a long-term fellowship from the Human Frontier Science Program (to A.E.X.B.).
Author information
Authors and Affiliations
Contributions
E.Y. created the WT2 system (hardware and software), analyzed data and wrote the paper. T.J. developed the WT2 analysis pipeline and built the online database. L.J.G. conducted all experiments. A.E.X.B. analyzed data, supervised research and wrote the paper. W.R.S. designed experiments, supervised research and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Note (PDF 2058 kb)
Supplementary Table 1
Supplementary Data Set (XLSX 19 kb)
Supplementary Table 2
Supplementary Data Set (XLSX 45 kb)
Supplementary Table 3
Supplementary Data Set (XLSX 48 kb)
Supplementary Table 4
Supplementary Data Set (XLSX 42 kb)
Supplementary Table 5
Supplementary Data Set (XLSX 6853 kb)
Rights and permissions
About this article
Cite this article
Yemini, E., Jucikas, T., Grundy, L. et al. A database of Caenorhabditis elegans behavioral phenotypes. Nat Methods 10, 877–879 (2013). https://doi.org/10.1038/nmeth.2560
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2560
This article is cited by
-
Automated recognition and analysis of body bending behavior in C. elegans
BMC Bioinformatics (2023)
-
Parallel pathways for serotonin biosynthesis and metabolism in C. elegans
Nature Chemical Biology (2023)
-
WormSwin: Instance segmentation of C. elegans using vision transformer
Scientific Reports (2023)
-
Multisite regulation integrates multimodal context in sensory circuits to control persistent behavioral states in C. elegans
Nature Communications (2023)
-
Long-term imaging reveals behavioral plasticity during C. elegans dauer exit
BMC Biology (2022)