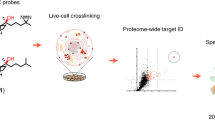
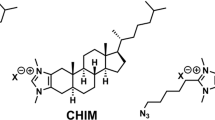
Abstract
Cholesterol is an essential structural component of cellular membranes and serves as a precursor for several classes of signaling molecules. Cholesterol exerts its effects and is, itself, regulated in large part by engagement in specific interactions with proteins. The full complement of sterol-binding proteins that exist in mammalian cells, however, remains unknown. Here we describe a chemoproteomic strategy that uses clickable, photoreactive sterol probes in combination with quantitative mass spectrometry to globally map cholesterol-protein interactions directly in living cells. We identified over 250 cholesterol-binding proteins, including receptors, channels and enzymes involved in many established and previously unreported interactions. Prominent among the newly identified interacting proteins were enzymes that regulate sugars, glycerolipids and cholesterol itself as well as proteins involved in vesicular transport and protein glycosylation and degradation, pointing to key nodes in biochemical pathways that may couple sterol concentrations to the control of other metabolites and protein localization and modification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schroeder, F. et al. Caveolin, sterol carrier protein-2, membrane cholesterol-rich microdomains and intracellular cholesterol trafficking. Subcell. Biochem. 51, 279–318 (2010).
Russell, D.W. Oxysterol biosynthetic enzymes. Biochim. Biophys. Acta 1529, 126–135 (2000).
McLean, K.J., Hans, M. & Munro, A.W. Cholesterol, an essential molecule: diverse roles involving cytochrome P450 enzymes. Biochem. Soc. Trans. 40, 587–593 (2012).
Russell, D.W. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50 (suppl.), S120–S125 (2009).
Badimon, L. & Vilahur, G. LDL-cholesterol versus HDL-cholesterol in the atherosclerotic plaque: inflammatory resolution versus thrombotic chaos. Ann. NY Acad. Sci. 1254, 18–32 (2012).
Mirza, R. et al. DHCR24 gene knockout mice demonstrate lethal dermopathy with differentiation and maturation defects in the epidermis. J. Invest. Dermatol. 126, 638–647 (2006).
Porter, F.D. Human malformation syndromes due to inborn errors of cholesterol synthesis. Curr. Opin. Pediatr. 15, 607–613 (2003).
Herman, G.E. Disorders of cholesterol biosynthesis: prototypic metabolic malformation syndromes. Hum. Mol. Genet. 12 Spec No 1, R75–R88 (2003).
Cianciola, N.L., Carlin, C.R. & Kelley, T.J. Molecular pathways for intracellular cholesterol accumulation: common pathogenic mechanisms in Niemann-Pick disease Type C and cystic fibrosis. Arch. Biochem. Biophys. 515, 54–63 (2011).
Rosenbaum, A.I. & Maxfield, F.R. Niemann-Pick type C disease: molecular mechanisms and potential therapeutic approaches. J. Neurochem. 116, 789–795 (2011).
Weinhofer, I., Forss-Petter, S., Kunze, M., Zigman, M. & Berger, J. X-linked adrenoleukodystrophy mice demonstrate abnormalities in cholesterol metabolism. FEBS Lett. 579, 5512–5516 (2005).
Brown, M.S. & Goldstein, J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997).
Gill, S., Chow, R. & Brown, A.J. Sterol regulators of cholesterol homeostasis and beyond: the oxysterol hypothesis revisited and revised. Prog. Lipid Res. 47, 391–404 (2008).
Jeon, T.I. & Osborne, T.F. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 (2012).
Heal, W.P., Jovanovic, B., Bessin, S., Wright, M.H., Magee, A.I. & Tate, E.W. Bioorthogonal chemical tagging of protein cholesterylation in living cells. Chem. Commun. (Camb.) 47, 4081–4083 (2011).
Hoop, C.L., Sivanandam, V.N., Kodali, R., Srnec, M.N. & van der Wel, P.C. Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry 51, 90–99 (2012).
Theesfeld, C.L., Pourmand, D., Davis, T., Garza, R.M. & Hampton, R.Y. The sterol-sensing domain (SSD) directly mediates signal-regulated endoplasmic reticulum-associated degradation (ERAD) of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase isozyme Hmg2. J. Biol. Chem. 286, 26298–26307 (2011).
Motamed, M. et al. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J. Biol. Chem. 286, 18002–18012 (2011).
Thiele, C., Hannah, M.J., Fahrenholz, F. & Huttner, W.B. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat. Cell Biol. 2, 42–49 (2000).
Charrin, S. et al. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33, 2479–2489 (2003).
Rostovtsev, V.V., Green, J.G., Fokin, V.V. & Sharpless, K.B. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Edn. Engl. 41, 2596–2599 (2002).
Speers, A.E. & Cravatt, B.F. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 11, 535–546 (2004).
Mintzer, E.A., Waarts, B.L., Wilschut, J. & Bittman, R. Behavior of a photoactivatable analog of cholesterol, 6-photocholesterol, in model membranes. FEBS Lett. 510, 181–184 (2002).
Kwon, H.J. et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213–1224 (2009).
Roberts, J.D. & Caserio, M. Basic Principles of Organic Chemistry 445–487 (Addison-Wesley, 1977).
Ong, S.E., Foster, L.J. & Mann, M. Mass spectrometric-based approaches in quantitative proteomics. Methods 29, 124–130 (2003).
Tong, F. et al. Decreased expression of ARV1 results in cholesterol retention in the endoplasmic reticulum and abnormal bile acid metabolism. J. Biol. Chem. 285, 33632–33641 (2010).
Thompson, A.A. et al. GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods 55, 310–317 (2011).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Pastorino, J.G. & Hoek, J.B. Regulation of hexokinase binding to VDAC. J. Bioenerg. Biomembr. 40, 171–182 (2008).
Christian, A.E., Haynes, M.P., Phillips, M.C. & Rothblat, G.H. Use of cyclodextrins for manipulating cellular cholesterol content. J. Lipid Res. 38, 2264–2272 (1997).
Martin, B.R., Wang, C., Adibekian, A., Tully, S.E. & Cravatt, B.F. Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84–89 (2012).
Washburn, M.P., Wolters, D. & Yates, J.R. III. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001).
Eng, J., McCormack, A.L. & Yates, J.R. An approach to correlate MS/MS data to amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010).
Acknowledgements
We thank P. Baran for helpful advice on the synthesis of sterol probes, and A. Rheingold and C. Moore for X-ray crystallographic analysis. This work was supported by US National Institutes of Health (CA132630) and the Skaggs Institute for Chemical Biology.
Author information
Authors and Affiliations
Contributions
J.J.H., S.E.T., M.J.N. and B.F.C. designed experiments; J.J.H., A.B.C. and S.E.T. performed experiments; J.J.H. and B.F.C. analyzed data; J.J.H. and B.F.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Table 5, Supplementary Note (PDF 2876 kb)
Supplementary Table 1
List of group I–IV proteins with experimental SILAC ratios. Groups I, II, III, and IV are presented on separate sheets, with the mean SILAC ratio obtained in the trans versus no–UV light, trans versus PEA-DEA, and trans versus 10 × cholesterol competition (Ch) shown. For proteins that were not detected in the no–UV light experiments but were found in the vehicle control experiments, the vehicle ratios are shown instead (colored in blue). For group I proteins, gene expression data are presented as 1 = yes; 0 = no, where ‘yes’ means that the gene expression change was > twofold at 30 min cholesterol treatment or > twofold at 12 h cholesterol treatment. All quantifications were manually filtered here. ND, not detected. See Supplementary Table 2 for a list of individual peptide sequences and ratios detected for each protein. (XLSX 152 kb)
Supplementary Table 2
Complete proteomic data for trans versus no UV light and versus vehicle. Data sets include the individual peptides detected for each protein, their SILAC ratios and charge states. SILAC ratios are presented as the mean of duplicates but are not manually filtered. (XLSX 1574 kb)
Supplementary Table 3
Data sets for each set of SILAC experiments. Mean ratios are presented on separate sheets for each set of SILAC experiments. Control experiments (no UV light and vehicle comparisons) are shown together, are unfiltered and were each run in duplicate. PEA-DA comparisons and cholesterol competition, shown together, were run in duplicate and quadruplicate, respectively, and background proteins from control runs are removed. Probe comparisons (trans versus epi, cis and trans), shown together, were run in duplicate, and background proteins are removed. All quantifications are raw and were not manually filtered. (XLSX 211 kb)
Supplementary Table 4
Complete gene expression profiles for cholesterol and probe treatments. The signal intensity for each gene in each microarray experiment is included. The three conditions tested were: (i) control/vehicle: cells were untreated, and were collected and processed in parallel with the other conditions; (ii) 30 min: cells received 20 μM cis probe + 100 μM cholesterol for 30 min before harvest; (iii) 12 h: cells received 20 μM cis probe + 100 μM cholesterol for 12 h before harvest. Total mRNA was extracted for each condition, and gene expression was analyzed by HU133 Set GeneChip from Affymetrix. (XLSX 6362 kb)
Supplementary Table 6
Unenriched versus trans-enrichment membrane proteome comparison by spectral counting. Raw data set presenting proteins in groups I–IV that were detected with >10 average spectral counts over quadruplicate trans probe–mediated enrichments (columns t1–t4) or unenriched membrane proteomes (columns u1–u4). (XLSX 108 kb)
Rights and permissions
About this article
Cite this article
Hulce, J., Cognetta, A., Niphakis, M. et al. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods 10, 259–264 (2013). https://doi.org/10.1038/nmeth.2368
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2368
This article is cited by
-
Enhanced mapping of small-molecule binding sites in cells
Nature Chemical Biology (2024)
-
DHCR24 reverses Alzheimer’s disease-related pathology and cognitive impairment via increasing hippocampal cholesterol levels in 5xFAD mice
Acta Neuropathologica Communications (2023)
-
Transcriptome sequencing and metabolome analysis reveal the metabolic reprogramming of partial hepatectomy and extended hepatectomy
BMC Genomics (2023)
-
Chemoproteomic mapping of the glycolytic targetome in cancer cells
Nature Chemical Biology (2023)
-
Identification of photocrosslinking peptide ligands by mRNA display
Communications Chemistry (2023)