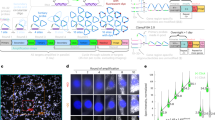
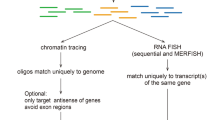
Abstract
We developed a cost-effective genome-scale PCR-based method for high-definition DNA FISH (HD-FISH). We visualized gene loci with diffraction-limited resolution, chromosomes as spot clusters and single genes together with transcripts by combining HD-FISH with single-molecule RNA FISH. We provide a database of over 4.3 million primer pairs targeting the human and mouse genomes that is readily usable for rapid and flexible generation of probes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Springer Protocols. Fluorescence In Situ Hybridization (FISH)–Application Guide (ed. Liehr, T.) (Springer, 2008).
Springer Protocols. Fluorescence In Situ Hybridization (FISH): Protocols and Applications (Methods in Molecular Biology). (eds Bridger, J.M. & Volpi, E.V.) (Humana Press, 2010).
Boyle, S., Rodesch, M.J., Halvensleben, H.A., Jeddeloh, J.A. & Bickmore, W.A. Chromosome Res. 19, 901–909 (2011).
Yamada, N.A. et al. Cytogenet. Genome Res. 132, 248–254 (2011).
Rogan, P.K., Cazcarro, P.M. & Knoll, J.H. Genome Res. 11, 1086–1094 (2001).
Navin, N. et al. Bioinformatics 22, 2437–2438 (2006).
Wiegant, J.C. et al. Cytogenet. Cell Genet. 87, 47–52 (1999).
Solovei, I. & Cremer, M. Methods Mol. Biol. 659, 117–126 (2010).
Cremer, T. & Cremer, M. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010).
Raj, A., van den Bogaard, P., Rifkin, S.A., van Oudenaarden, A. & Tyagi, S. Nat. Methods 5, 877–879 (2008).
Tsafrir, D. et al. Cancer Res. 66, 2129–2137 (2006).
Beliveau, B.J. et al. Proc. Natl. Acad. Sci. USA published online, doi:10.1073/pnas.1213818110 (2012).
Kent, W.J. Genome Res. 12, 656–664 (2002).
Untergasser, A. et al. Nucleic Acids Res. 40, e115 (2012).
Wheeler, D.L. et al. Nucleic Acids Res. 31, 28–33 (2003).
Itzkovitz, S. et al. Nat. Cell Biol. 14, 106–114 (2012).
Acknowledgements
We thank P. Junker and S. Semrau for helpful discussions. We are grateful to R.A. Weinberg (Massachusetts Institute of Technology) for providing hTERT-HME1 cells. This work was supported by the US National Institutes of Health (NIH)/National Cancer Institute Physical Sciences Oncology Center at Massachusetts Institute of Technology (U54CA143874), an NIH Pioneer award (1DP1OD003936) and a Nederlandse Organisatie voor Wetenschappelijk Onderzoek Vici award to A.v.O. M.B. and S.I. are sponsored by the Human Frontiers Science Program.
Author information
Authors and Affiliations
Contributions
N.C. and A.v.O. conceived the methods. M.B. and N.C. performed experiments, analyzed the data and wrote the manuscript. L.T. generated the genome-wide primer databases, designed the probes and wrote the manuscript. S.K. and S.I. developed software for image processing, provided suggestions on data analysis and corrected the manuscript. A.v.O. guided experiments and data analysis, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Note (PDF 9821 kb)
3D rendering of Chr17 in HME cells, visualized with ten HD-FISH probes evenly spaced every 8 Mb and labeled with two alternating fluorophores (green: AlexaFluor594; magenta: AlexaFluor647).
The nucleus displayed is the same as in the Z-projection shown in Figure 3a (mid panel). (MOV 5461 kb)
41592_2013_BFnmeth2306_MOESM170_ESM.mov
3D animation of Chr17 in HME cells, visualized with sixteen HD-FISH probes spaced evenly every 5 Mb and labeled with two alternating fluorophores (green: AlexaFluor594; magenta: AlexaFluor647) together with a Chr17 paint probe (blue). (MOV 4960 kb)
Rights and permissions
About this article
Cite this article
Bienko, M., Crosetto, N., Teytelman, L. et al. A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat Methods 10, 122–124 (2013). https://doi.org/10.1038/nmeth.2306
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2306
This article is cited by
-
Chromosome-level assembly of the Rangifer tarandus genome and validation of cervid and bovid evolution insights
BMC Genomics (2023)
-
Spatial organization of transcribed eukaryotic genes
Nature Cell Biology (2022)
-
PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments
Nature Methods (2021)
-
Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M
Nature Protocols (2020)
-
iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture
Nature Communications (2019)