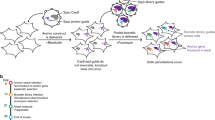
Abstract
Knockout collections are invaluable tools for studying model organisms such as yeast. However, there are no large-scale knockout collections of human cells. Using gene-trap mutagenesis in near-haploid human cells, we established a platform to generate and isolate individual 'gene-trapped cells' and used it to prepare a collection of human cell lines carrying single gene-trap insertions. In most cases, the insertion can be reversed. This growing library covers 3,396 genes, one-third of the expressed genome, is DNA-barcoded and allows systematic screens for a wide variety of cellular phenotypes. We examined cellular responses to TNF-α, TGF-β, IFN-γ and TNF-related apoptosis-inducing ligand (TRAIL), to illustrate the value of this unique collection of isogenic human cell lines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Subtelny, S. The development of haploid and homozygous diploid frog embryos obtained from transplantations of haploid nuclei. J. Exp. Zool. 139, 263–305 (1958).
Thorne, M.H., Collins, R.K. & Sheldon, B.L. Live haploid-diploid and other unusual mosaic chickens (Gallus domesticus). Cytogenet. Cell Genet. 45, 21–25 (1987).
Corley-Smith, G.E., Lim, C.J. & Brandhorst, B.P. Production of androgenetic zebrafish (Danio rerio). Genetics 142, 1265–1276 (1996).
Yi, M., Hong, N. & Hong, Y. Generation of medaka fish haploid embryonic stem cells. Science 326, 430–433 (2009).
Elling, U. et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell 9, 563–574 (2011).
Leeb, M. & Wutz, A. Derivation of haploid embryonic stem cells from mouse embryos. Nature 479, 131–134 (2011).
Kotecki, M., Reddy, P.S. & Cochran, B.H. Isolation and characterization of a near-haploid human cell line. Exp Cell Res 252, 273–280 (1999).
Andersson, B.S. et al. Ph-positive chronic myeloid leukemia with near-haploid conversion in vivo and establishment of a continuously growing cell line with similar cytogenetic pattern. Cancer Genet. Cytogenet. 24, 335–343 (1987).
Holmfeldt, K., Odic, D., Sullivan, M.B., Middelboe, M. & Riemann, L. Cultivated single-stranded DNA phages that infect marine Bacteroidetes prove difficult to detect with DNA-binding stains. Appl. Environ. Microbiol. 78, 892–894 (2012).
Carette, J.E. et al. Generation of iPSCs from cultured human malignant cells. Blood 115, 4039–4042 (2010).
Carette, J.E. et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235 (2009).
Carette, J.E. et al. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 29, 542–546 (2011).
Carette, J.E. et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011).
Guimaraes, C.P. et al. Identification of host cell factors required for intoxication through use of modified cholera toxin. J. Cell Biol. 195, 751–764 (2011).
Papatheodorou, P. et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc. Natl. Acad. Sci. USA 108, 16422–16427 (2011).
Reiling, J.H. et al. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc. Natl. Acad. Sci. USA 108, 11756–11765 (2011).
Rosmarin, D.M. et al. Attachment of Chlamydia trachomatis L2 to host cells requires sulfation. Proc. Natl. Acad. Sci. USA 109, 10059–10064 (2012).
Birsoy, K. et al. MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat. Genet. 45, 104–108 (2013).
Jacobson, L.S. et al. Cathepsin-mediated necrosis controls the adaptive immune response by Th2 (T helper type 2)-associated adjuvants. J. Biol. Chem. 288, 7481–7491 (2013).
Shi, Q. & King, R.W. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 437, 1038–1042 (2005).
Lundberg, E. et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450 (2010).
Lewinski, M.K. et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2, e60 (2006).
Scheinecker, C. & Smolen, J.S. Rheumatoid arthritis in 2010: from the gut to the joint. Nat. Rev. Rheumatol. 7, 73–75 (2011).
Moustakas, A. & Heldin, C.H. The regulation of TGFbeta signal transduction. Development 136, 3699–3714 (2009).
Varfolomeev, E.E. et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9, 267–276 (1998).
Kischkel, F.C. et al. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 12, 611–620 (2000).
Biankin, A.V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Manceau, G. et al. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma. J. Int. Cancer 132, 2217–2221 (2013).
Hodis, E. et al. A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012).
Li, M. et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011).
Yan, Z. et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 19, 1662–1667 (2005).
Orlean, P. & Menon, A.K. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48, 993–1011 (2007).
Skarnes, W.C. et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 36, 543–544 (2004).
Stanford, W.L., Cohn, J.B. & Cordes, S.P. Gene-trap mutagenesis: past, present and beyond. Nat. Rev. Genet. 2, 756–768 (2001).
Fimia, G.M. et al. Ambra1 regulates autophagy and development of the nervous system. Nature 447, 1121–1125 (2007).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Wisniewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Bennett, K.L. et al. Proteomic analysis of human cataract aqueous humour: Comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ(R)-labelled specimens. J. Proteomics 74, 151–166 (2011).
Colinge, J., Masselot, A., Giron, M., Dessingy, T. & Magnin, J. OLAV: towards high-throughput tandem mass spectrometry data identification. Proteomics 3, 1454–1463 (2003).
Geiger, T., Wehner, A., Schaab, C., Cox, J. & Mann, M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell Proteomics 11, M111 014050 (2012).
Acknowledgements
We thank G. Winter and members of the Brummelkamp, Nijman and Superti-Furga laboratories for discussions and technical assistance, R. Martins for help with the illustrations in Figure 2, O. Majdic (Medical University of Vienna) for providing antibodies, J. Carette for advice on gene trap vector design, and H. Pickersgill for manuscript editing and suggestions. C. Banning was supported by a FemPower grant from Zentrum für Innovation und Technologie (Die Technologieagentur der Stadt Wien), and A.L. and M.L. were supported by a Zentrum für Innovation und Technologie Life Sciences 2011 grant. M.R. was supported by European Molecular Biology Organization fellowship (ALTF1346-2011). K.P. was supported by a European Research Council grant (ERC-2009-AdG-250179-i-FIVE).
Author information
Authors and Affiliations
Contributions
S.M.B.N., T.R.B. and G.S.-F. conceived the haploid gene-trap mutant collection and provided overall guidance. S.M.B.N. and T.B. analyzed data and, together with G.S.-F. and T.R.B., wrote the paper. S.M.B.N., T.R.B., T.B. and G.C. conceived the gene-trap vector design including barcodes and loxP sites and the clone-mapping pipeline. T.B. and C. Banning supervised the establishment of the mutant collection and performed validation experiments. P.H., A.L., M.L., W.F., S.S. and M.R.T.C. assisted in the establishment of the mutant collection and validation experiments. F.M.P., D.C., N.T., F.S., B.E., P.M.G., V.A.B., T.K., B.G., C. Bock and R.K. generated the samples for DNA and RNA sequencing and SNP arrays, and analyzed the data. R.S., B.E. and G.C. established the clone-mapping bioinformatics pipeline and databases. C.K., M.R., M.S. and F.F.d.l.C. performed clone-validation experiments. K.P., K.L.B. and J.C. generated samples for mass spectrometry and analyzed the data. B.M., J.S. and S.K. performed and analyzed the leukocyte typing experiments. G.C. and G.A. assisted in platform design.
Corresponding authors
Ethics declarations
Competing interests
G.A., G.C., T.R.B., G.S.-F. and S.M.B.N. are founders and shareholders of Haplogen. T.B., C.Banning, P.H., A.L., M.L., W.F., S.S. and M.R.T.C. are employees of Haplogen.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1, 2, 7, 9 and 11–14 (PDF 7324 kb)
Supplementary Table 3
Variants from exome and whole genome (100-bp paired-end) sequencing. (XLSX 23712 kb)
Supplementary Table 4
Comprehensive proteome profiling. (XLSX 1742 kb)
Supplementary Table 5
mRNA and protein expression. (XLSX 426 kb)
Supplementary Table 6
KEGG pathway analysis of gene trap collection. (XLSX 21 kb)
Supplementary Table 8
KEGG pathway coverage of clone collection. (XLS 53 kb)
Supplementary Table 10
Detailed information concerning all available gene trap clones. (XLS 1802 kb)
Source data
Rights and permissions
About this article
Cite this article
Bürckstümmer, T., Banning, C., Hainzl, P. et al. A reversible gene trap collection empowers haploid genetics in human cells. Nat Methods 10, 965–971 (2013). https://doi.org/10.1038/nmeth.2609
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2609
This article is cited by
-
Evidence for widespread existence of functional novel and non-canonical human transcripts
BMC Biology (2023)
-
CATS: Cas9-assisted tag switching. A high-throughput method for exchanging genomic peptide tags in yeast
BMC Genomics (2020)
-
Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import
Nature Communications (2020)
-
The essentiality landscape of cell cycle related genes in human pluripotent and cancer cells
Cell Division (2019)
-
High-throughput genome-wide phenotypic screening via immunomagnetic cell sorting
Nature Biomedical Engineering (2019)