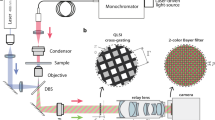
Abstract
Conventional acquisition of three-dimensional (3D) microscopy data requires sequential z scanning and is often too slow to capture biological events. We report an aberration-corrected multifocus microscopy method capable of producing an instant focal stack of nine 2D images. Appended to an epifluorescence microscope, the multifocus system enables high-resolution 3D imaging in multiple colors with single-molecule sensitivity, at speeds limited by the camera readout time of a single image.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fischer, R.S., Wu, Y., Kanchanawong, P., Shroff, H. & Waterman, C.M. Trends Cell Biol. 21, 682–691 (2011).
Botcherby, E.J., Juskaitis, R., Booth, M.J. & Wilson, T. Opt. Lett. 32, 2007–2009 (2007).
Prabhat, P., Ram, S., Ward, E.S. & Ober, R.J. IEEE Trans. Nanobioscience 3, 237–242 (2004).
Blanchard, P.M. & Greenaway, A.H. Appl. Opt. 38, 6692–6699 (1999).
Blanchard, P.M. & Greenaway, A.H. Opt. Commun. 183, 29–36 (2000).
Mait, J.N. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 12, 2145–2158 (1995).
Born, M. & Wolf, E. Principles of Optics (Cambridge University Press, 1999).
Lawrimore, J., Bloom, K.S. & Salmon, E.D. J. Cell Biol. 195, 573–582 (2011).
Coffman, V.C., Wu, P., Parthun, M.R. & Wu, J.Q. J. Cell Biol. 195, 563–572 (2011).
Pearson, C.G., Maddox, P.S., Salmon, E.D. & Bloom, K. J. Cell Biol. 152, 1255–1266 (2001).
Pinaud, F., Clarke, S., Sittner, A. & Dahan, M. Nat. Methods 7, 275–285 (2010).
Darzacq, X. et al. Nat. Struct. Mol. Biol. 14, 796–806 (2007).
Pavani, S.R. et al. Proc. Natl. Acad. Sci. USA 106, 2995–2999 (2009).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Science 319, 810–813 (2008).
Ram, S., Prabhat, P., Chao, J., Ward, E.S. & Ober, R.J. Biophys. J. 95, 6025–6043 (2008).
Wu, Y. et al. Proc. Natl. Acad. Sci. USA 108, 17708–17713 (2011).
Neil, M.A., Wilson, T. & Juskaitis, R. J. Microsc. 197, 219–223 (2000).
Goodman, J.W. Introduction to Fourier Optics (Roberts & Co., 2005).
Jaqaman, K. et al. Nat. Methods 5, 695–702 (2008).
Acknowledgements
We dedicate this paper to the late Mats Gustafsson. We thank K. Wicker, R. Heinzmann, T. Wilson and D. Grunwald for valuable discussions about multifocus microscopy; C. Zimmer for his help with the 3D localization algorithm; F. Amat and A. Verma for assistance with data processing; L. Shao, L. Winoto, J. Sedat, R. Singer and E. Betzig for discussions on 3D microscopy; V. Iyer for discussions on hardware control; Z. Zhang and Y. Li (HHMI Janelia Farm) for sharing reagents; H. White and H. Rego for assistance in live-cell imaging; and V. Butler and R. Kerr (HHMI Janelia Farm) and W. Schafer (MRC Laboratory of Molecular Biology) for the C. elegans strain. S.A. would like to thank G. Rubin, K. Moses and R. Tijan for ensuring continued funding for this project during and after the difficult time of M.G.'s sickness. A.Y.K. is supported by a Jane Coffin Childs postdoctoral fellowship. M.D. acknowledges the support of a Fulbright fellowship. X.D. and M.D. are supported by grant ANR-08-PCVI-0013 from Agence Nationale pour la Recherche. D.A.A. is supported by US National Institutes of Health grant GM31627. S.A., C.I.B. and D.A.A. are supported by the HHMI. J.W., G.M. and C.W. are supported by the Center for Cancer Research, National Cancer Institute and HHMI.
Author information
Authors and Affiliations
Contributions
The optical layout was conceived by M.G.L.G. Optical design and optimizations were made by M.G.L.G. and S.A. S.A. built the system and implemented the hardware control electronics. J.C., B.H. and M.D. performed single-molecule and yeast experiments. S.A., B.H., J.C. and M.D. developed image processing tools, and B.H. and J.C. analyzed the data. S.A. and A.Y.K. acquired the C. elegans data. J.W. and G.M. constructed the yeast strain; J.W. and C.W. participated in centromere imaging. P.S., C.D.D. and X.D. constructed and characterized the RPB1 cellular system. S.S. contributed to the theoretical performance evaluation of the microscope. F.M. and X.D. provided the 3D single-emitter detection algorithm. M.G.L.G., D.A.A. and C.I.B. supervised the project. S.A. and M.D. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Notes 1–4 (PDF 6953 kb)
41592_2013_BFnmeth2277_MOESM595_ESM.mov
Raw data of 200-nm beads mounted on a coverslip and displaced along the z axis in 100-nm steps using a piezoelectric stage. (MOV 3080 kb)
41592_2013_BFnmeth2277_MOESM596_ESM.avi
The reconstructed 3D rendered movie of the data stack of Supplementary Video 1, showing the beads realigned inside the imaging volume. (AVI 1649 kb)
Raw data of yeast cells expressing Cse4-GFP.
Lateral field of view 20×20 μm. Focus step between successive planes Δz = 380 nm. (Voxel size: x,y = 120 nm, z = 380 nm.) Data were acquired at a speed of one multifocus image (exposure time 100 ms) every 3 s. Scale bar, 1.5 μm. (AVI 19340 kb)
3D reconstruction of Supplementary Video 3, visualizing the 3D dynamics of yeast centromeres during anaphase.
Upper movie, xy view; lower movie, xz view. Scale bar, 1 μm. (AVI 20190 kb)
3D view of single trajectories of RNA polymerase II (magenta) diffusing in the nucleus of U2OS cells transfected with lamin B1–GFP as a nuclear membrane marker (green).
Upper movie, xy view; lower movie, xz view. Only trajectories longer than five frames are displayed as segments. Data were acquired at a speed of 35 volumes s–1. (Voxel size: x,y = 120 nm, z = 380 nm.) (AVI 17620 kb)
Examples of 3D trajectories, exhibiting slow diffusion, rapid diffusion and mixed behavior.
Data were acquired at a speed of 35 volumes s–1. (AVI 7399 kb)
The unc-47 GABAergic motor neurons of this C. elegans embryo express a green fluorescent protein.
This movie shows the raw multifocus data, a set of nine simultaneously formed focal planes (60×60 μm at 2-μm separation covering a depth of 18 μm) arranged in a 3×3 array on the camera. Data were recorded at 9 Hz for 5.5 min and are displayed at 10× this speed. Scale bar, 20 μm. (AVI 2867 kb)
41592_2013_BFnmeth2277_MOESM602_ESM.avi
Average-intensity projection along z of the 3D assembled data of the C. elegans embryo in Supplementary Video 7. (AVI 1842 kb)
Rights and permissions
About this article
Cite this article
Abrahamsson, S., Chen, J., Hajj, B. et al. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat Methods 10, 60–63 (2013). https://doi.org/10.1038/nmeth.2277
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2277
This article is cited by
-
Fast topographic optical imaging using encoded search focal scan
Nature Communications (2024)
-
High-fidelity 3D live-cell nanoscopy through data-driven enhanced super-resolution radial fluctuation
Nature Methods (2023)
-
Rapid detection of neurons in widefield calcium imaging datasets after training with synthetic data
Nature Methods (2023)
-
Optical-resolution photoacoustic microscopy with a needle-shaped beam
Nature Photonics (2023)
-
Imagining the future of optical microscopy: everything, everywhere, all at once
Communications Biology (2023)