Abstract
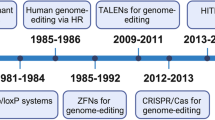
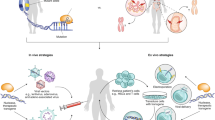
Recent progress in developing programmable nucleases, such as zinc-finger nucleases, transcription activator–like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeat (CRISPR)–Cas nucleases, have paved the way for gene editing to enter clinical practice. This translation is a result of combining high nuclease activity with high specificity and successfully applying this technology in various preclinical disease models, including infectious disease, primary immunodeficiencies, hemoglobinopathies, hemophilia and muscular dystrophy. Several clinical gene-editing trials, both ex vivo and in vivo, have been initiated in the past 2 years, including studies that aim to knockout genes as well as to add therapeutic transgenes. Here we discuss the advances made in the gene-editing field in recent years, and specify priorities that need to be addressed to expand therapeutic genome editing to further disease entities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 109, E2579–E2586 (2012).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cox, D.B., Platt, R.J. & Zhang, F. Therapeutic genome editing: prospects and challenges. Nat. Med. 21, 121–131 (2015).
Marketsandmarkets.comGenome Editing/Genome Engineering Market by Technology (CRISPR, TALEN, ZFN), Applications (Cell Line Engineering, Animal Genetic Engineering, Plant Genetic Engineering), End User (Biotechnology & Pharmaceutical Companies, CROs)—Global Forecast to 2021 (marketsandmarkets.com, 2016).
Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 188, 773–782 (2011).
Joung, J.K. & Sander, J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 (2013).
Naldini, L. Gene therapy returns to centre stage. Nature 526, 351–360 (2015).
Capecchi, M.R. Altering the genome by homologous recombination. Science 244, 1288–1292 (1989).
Rouet, P., Smih, F. & Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. USA 91, 6064–6068 (1994).
Urnov, F.D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005).
Hoban, M.D. et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, 2597–2604 (2015).
Genovese, P. et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240 (2014).
De Ravin, S.S. et al. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 34, 424–429 (2016).
Hubbard, N. et al. Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood 127, 2513–2522 (2016).
Traxler, E.A. et al. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 22, 987–990 (2016).
Dever, D.P. et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016).
Sather, B.D. et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 7, 307ra156 (2015).
Tebas, P. et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 370, 901–910 (2014).
Poirot, L. et al. Multiplex genome-edited T cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 75, 3853–3864 (2015).
Qasim, W. et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 9, eaaj2013 (2017).
Hoban, M.D. et al. CRISPR/Cas9-mediated correction of the sickle mutation in human CD34+ cells. Mol. Ther. 24, 1561–1569 (2016).
Chu, V.T. et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015).
Maruyama, T. et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33, 538–542 (2015).
Yu, C. et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16, 142–147 (2015).
Song, J. et al. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 7, 10548 (2016).
Kaufmann, K.B., Büning, H., Galy, A., Schambach, A. & Grez, M. Gene therapy on the move. EMBO Mol. Med. 5, 1642–1661 (2013).
Li, H. et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221 (2011).
Sharma, R. et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126, 1777–1784 (2015).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
Pankowicz, F.P. et al. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat. Commun. 7, 12642 (2016).
Nelson, C.E. et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407 (2016).
Tabebordbar, M. et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411 (2016).
Long, C. et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 (2016).
Williams, D.A. & Thrasher, A.J. Concise review: lessons learned from clinical trials of gene therapy in monogenic immunodeficiency diseases. Stem Cells Transl. Med. 3, 636–642 (2014).
Solinís, M.A., del Pozo-Rodríguez, A., Apaolaza, P.S. & Rodríguez-Gascón, A. Treatment of ocular disorders by gene therapy. Eur. J. Pharm. Biopharm. 95Pt B, 331–342 (2015).
Ruiz de Garibay, A.P., Delgado, D., Del Pozo-Rodríguez, A., Solinís, M.A. & Gascón, A.R. Multicomponent nanoparticles as nonviral vectors for the treatment of Fabry disease by gene therapy. Drug Des. Devel. Ther. 6, 303–310 (2012).
Yin, H. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328–333 (2016).
Zuris, J.A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2014).
Hornung, V. & Latz, E. Intracellular DNA recognition. Nat. Rev. Immunol. 10, 123–130 (2010).
Wang, J. et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 33, 1256–1263 (2015).
Holkers, M. et al. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods 11, 1051–1057 (2014).
Mock, U. et al. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 43, 5560–5571 (2015).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Hu, J.H., Davis, K.M. & Liu, D.R. Chemical biology approaches to genome editing: understanding, controlling, and delivering programmable nucleases. Cell Chem. Biol. 23, 57–73 (2016).
Kim, S., Kim, D., Cho, S.W., Kim, J. & Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Yu, K.R., Natanson, H. & Dunbar, C.E. Gene editing of human hematopoietic stem and progenitor cells: promise and potential hurdles. Hum. Gene Ther. 27, 729–740 (2016).
Doyon, Y. et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods 8, 74–79 (2011).
Söllü, C. et al. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 38, 8269–8276 (2010).
Szczepek, M. et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 25, 786–793 (2007).
Miller, J.C. et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25, 778–785 (2007).
Maeder, M.L. et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 31, 294–301 (2008).
Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang, H.S. & Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Miller, J.C. et al. Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat. Methods 12, 465–471 (2015).
Juillerat, A. et al. Optimized tuning of TALEN specificity using non-conventional RVDs. Sci. Rep. 5, 8150 (2015).
Guilinger, J.P. et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods 11, 429–435 (2014).
Cho, S.W., Kim, S., Kim, J.M. & Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Cradick, T.J., Fine, E.J., Antico, C.J. & Bao, G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 41, 9584–9592 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013).
Mali, P., Esvelt, K.M. & Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013).
Tsai, S.Q. & Joung, J.K. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 17, 300–312 (2016).
Slaymaker, I.M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Kleinstiver, B.P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Ran, F.A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Müller, M. et al. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol. Ther. 24, 636–644 (2016).
Lee, C.M., Cradick, T.J. & Bao, G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol. Ther. 24, 645–654 (2016).
Kleinstiver, B.P. et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Ramirez, C.L. et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 40, 5560–5568 (2012).
Ran, F.A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Luo, Y. et al. Generation of TALE nickase-mediated gene-targeted cows expressing human serum albumin in mammary glands. Sci. Rep. 6, 20657 (2016).
Komor, A.C., Kim, Y.B., Packer, M.S., Zuris, J.A. & Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439 (2014).
Sander, J.D. & Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355 (2014).
Ousterout, D.G. et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 6, 6244 (2015).
Amabile, A. et al. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 167, 219–232.e214 (2016).
Tsai, S.Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Lee, C.M., Cradick, T.J., Fine, E.J. & Bao, G. Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol. Ther. 24, 475–487 (2016).
Baum, C. et al. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol. Ther. 9, 5–13 (2004).
Mussolino, C. et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–9293 (2011).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Mussolino, C. & Cathomen, T. TALE nucleases: tailored genome engineering made easy. Curr. Opin. Biotechnol. 23, 644–650 (2012).
Cho, S.W. et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24, 132–141 (2014).
Tsai, S.Q. et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32, 569–576 (2014).
Canver, M.C. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015).
Avalle, L., Pensa, S., Regis, G., Novelli, F. & Poli, V. STAT1 and STAT3 in tumorigenesis: A matter of balance. JAK-STAT 1, 65–72 (2012).
Pike-Overzet, K. et al. Successful RAG1-SCID gene therapy depends on the level of RAG1 expression. J. Allergy Clin. Immunol. 134, 242–243 (2014).
Ding, W. et al. Zinc finger nucleases targeting the human papillomavirus E7 oncogene induce E7 disruption and a transformed phenotype in HPV16/18-positive cervical cancer cells. Clin. Cancer Res. 20, 6495–6503 (2014).
Bloom, K., Ely, A., Mussolino, C., Cathomen, T. & Arbuthnot, P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol. Ther. 21, 1889–1897 (2013).
Holt, N. et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 28, 839–847 (2010).
Perez, E.E. et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 26, 808–816 (2008).
Su, S. et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 6, 20070 (2016).
Valton, J. et al. A multidrug-resistant engineered CAR T cell for allogeneic combination immunotherapy. Mol. Ther. 23, 1507–1518 (2015).
Provasi, E. et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 18, 807–815 (2012).
Lombardo, A. et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25, 1298–1306 (2007).
Yang, Y. et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 34, 334–338 (2016).
Acknowledgements
We thank all members of the laboratories for helpful discussion. Current research in our labs is funded by the Federal Ministry of Education and Research (IFB—01EO0803; iMACnet—01EK1602B; HBV-TALE—01DG15005), the German Research Foundation (SFB1160—TP17), the European Commission's Horizon 2020 Program (SCIDNET—666908; CARAT—667980), Casebia Therapeutics (CAU-950-20140702) and the German Academic Exchange Service.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
T.C. is a consultant for TRACR Hematology.
Rights and permissions
About this article
Cite this article
Cornu, T., Mussolino, C. & Cathomen, T. Refining strategies to translate genome editing to the clinic. Nat Med 23, 415–423 (2017). https://doi.org/10.1038/nm.4313
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4313
This article is cited by
-
Extend the benchmarking indel set by manual review using the individual cell line sequencing data from the Sequencing Quality Control 2 (SEQC2) project
Scientific Reports (2024)
-
Enhancement of the viability of T cells electroporated with DNA via osmotic dampening of the DNA-sensing cGAS–STING pathway
Nature Biomedical Engineering (2023)
-
Whole genomic analysis reveals atypical non-homologous off-target large structural variants induced by CRISPR-Cas9-mediated genome editing
Nature Communications (2023)
-
Interstrand crosslinking of homologous repair template DNA enhances gene editing in human cells
Nature Biotechnology (2023)
-
mRNA-based cancer therapeutics
Nature Reviews Cancer (2023)